+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of TIR-STING/c-di-GMP complex | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | NADase / HYDROLASE | |||||||||
| 機能・相同性 | CD-NTase-associated protein 12/Pycsar effector protein, TIR domain / CAP12/Pycsar effector protein, TIR domain / Prokaryotic STING domain / Prokaryotic STING domain / NAD+ glycohydrolase / NADP+ nucleosidase activity / defense response to virus / nucleotide binding / CD-NTase-associated protein 12 機能・相同性情報 機能・相同性情報 | |||||||||
| 生物種 |  Epilithonimonas lactis (バクテリア) Epilithonimonas lactis (バクテリア) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.88 Å | |||||||||
 データ登録者 データ登録者 | Lu DF / Liu S | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: mBio / 年: 2025 ジャーナル: mBio / 年: 2025タイトル: Structural insights into distinct filamentation states reveal a regulatory mechanism for bacterial STING activation. 著者: Yuchao Yang / Yueyue Liu / Xue Ma / Xuan Zhao / Jian Cao / Yu Liu / Shanqin Li / Jing Wu / Yuanzhu Gao / Lianwan Chen / Changxin Wu / Guijun Shang / Sheng Liu / Defen Lu /  要旨: The cyclic oligonucleotide-based antiphage signaling system (CBASS) is a bacterial immune mechanism that was evolutionarily linked to the eukaryotic cGAS-STING pathway, which protects against phage ...The cyclic oligonucleotide-based antiphage signaling system (CBASS) is a bacterial immune mechanism that was evolutionarily linked to the eukaryotic cGAS-STING pathway, which protects against phage infection through abortive cell death. CBASS operons encode cyclic dinucleotide synthases (CD-NTases) and effector proteins (Caps), such as bacterial STING, which senses cyclic dinucleotides like 3'3'-c-di-GMP to trigger defense. Although bacterial STING oligomerizes into filaments upon ligand binding, the functional roles of distinct filament states remain unclear. Here, we resolve cryo-EM structures of TIR-STING (STING) bound to 3'3'-c-di-GMP, revealing two oligomeric states: spiral-shaped single filaments and fiber bundles composed of straight protofibrils. In spiral filaments, the STING domain sequesters the TIR domain's BB loop within a hydrophobic core, suppressing NADase activity. This inactive conformation is stabilized by interactions between the CBDα4 helix and the TIR domain, as well as a calcium-binding site. Conversely, fiber bundle formation-driven by inter-protofibril TIR domain interactions-disrupts these autoinhibitory contacts, liberating the BB loop to enable head-to-tail assembly of adjacent TIR domains into a composite NADase-active site. Calcium ions promote spiral filament assembly while inhibiting fiber bundles, revealing a dual regulatory role in tuning STING activation. Strikingly, this mechanism diverges from single-filament systems like STING, underscoring evolutionary diversity in STING signaling. Our findings establish distinct filament architectures as structural checkpoints governing bacterial STING activation, providing mechanistic insights into how conformational plasticity and environmental cues like calcium regulate abortive infection. These results highlight parallels between prokaryotic and eukaryotic immune strategies, emphasizing conserved principles in pathogen defense across domains of life.IMPORTANCEBacteria employ a sophisticated immune system, CBASS, evolutionarily related to human antiviral pathways, to defend against viral (phage) attacks. This study reveals how the bacterial protein STING acts as a molecular switch, transitioning between an inactive spiral structure stabilized by calcium ions and an active fiber bundle. When calcium levels drop, STING reorganizes into fiber bundles, activating its ability to degrade essential cellular molecules. This self-destructive mechanism halts phage replication by sacrificing the infected cell, protecting the bacterial population. The findings demonstrate how structural rearrangements govern life-or-death immune decisions, mirroring principles in human STING signaling. By uncovering calcium's role in regulating this process, the work deepens our understanding of microbial immunity and highlights shared strategies across domains of life. These insights could inspire novel antimicrobial therapies or bioengineered systems to combat infections, bridging fundamental science with practical applications in health and biotechnology. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_63221.map.gz emd_63221.map.gz | 59.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-63221-v30.xml emd-63221-v30.xml emd-63221.xml emd-63221.xml | 15.4 KB 15.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_63221.png emd_63221.png | 40.5 KB | ||
| Filedesc metadata |  emd-63221.cif.gz emd-63221.cif.gz | 5.7 KB | ||
| その他 |  emd_63221_half_map_1.map.gz emd_63221_half_map_1.map.gz emd_63221_half_map_2.map.gz emd_63221_half_map_2.map.gz | 59 MB 59 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63221 http://ftp.pdbj.org/pub/emdb/structures/EMD-63221 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63221 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63221 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_63221_validation.pdf.gz emd_63221_validation.pdf.gz | 864.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_63221_full_validation.pdf.gz emd_63221_full_validation.pdf.gz | 863.6 KB | 表示 | |
| XML形式データ |  emd_63221_validation.xml.gz emd_63221_validation.xml.gz | 12.2 KB | 表示 | |
| CIF形式データ |  emd_63221_validation.cif.gz emd_63221_validation.cif.gz | 14.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63221 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63221 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63221 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-63221 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  9lmqMC  9lmrC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_63221.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_63221.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ボクセルのサイズ | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : TIR-STING/c-di-GMP complex
| 全体 | 名称: TIR-STING/c-di-GMP complex |
|---|---|
| 要素 |
|
-超分子 #1: TIR-STING/c-di-GMP complex
| 超分子 | 名称: TIR-STING/c-di-GMP complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  Epilithonimonas lactis (バクテリア) Epilithonimonas lactis (バクテリア) |
-分子 #1: CD-NTase-associated protein 12
| 分子 | 名称: CD-NTase-associated protein 12 / タイプ: protein_or_peptide / ID: 1 / コピー数: 8 / 光学異性体: LEVO / EC番号: NAD+ glycohydrolase |
|---|---|
| 由来(天然) | 生物種:  Epilithonimonas lactis (バクテリア) Epilithonimonas lactis (バクテリア) |
| 分子量 | 理論値: 35.717707 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MRTRIFIGSS KEGLKEANYV KSRLEKANFE VFIWNDDIFK PNKNTLETLL NVASLFDFGI MIATKDDFTA SRDDIFETVR DNVVFEFGL FLGRLGENRA FALQENGAKL PSDLLGITIP KFEKTDDYFS NYNLNTEIDN IIKIINEKIS LGELGLLPST V LAIGYYEN ...文字列: MRTRIFIGSS KEGLKEANYV KSRLEKANFE VFIWNDDIFK PNKNTLETLL NVASLFDFGI MIATKDDFTA SRDDIFETVR DNVVFEFGL FLGRLGENRA FALQENGAKL PSDLLGITIP KFEKTDDYFS NYNLNTEIDN IIKIINEKIS LGELGLLPST V LAIGYYEN FVSTVCDALH SLPTIKLNGI EYKDFVFNII IPNDLDADIK RRAQIYFKKM DIHEVKIDTN GRSFPLYLQI DE ENSGDVA VLYDMPTTLG GIDKAIEMYM KKGHIGKTSQ QQLLEERELR NFKTTLINLI NNNSFTKTFV KVIEE UniProtKB: CD-NTase-associated protein 12 |
-分子 #2: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydr...
| 分子 | 名称: 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one) タイプ: ligand / ID: 2 / コピー数: 4 / 式: C2E |
|---|---|
| 分子量 | 理論値: 690.411 Da |
| Chemical component information | 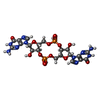 ChemComp-C2E: |
-分子 #3: CALCIUM ION
| 分子 | 名称: CALCIUM ION / タイプ: ligand / ID: 3 / コピー数: 4 / 式: CA |
|---|---|
| 分子量 | 理論値: 40.078 Da |
-分子 #4: water
| 分子 | 名称: water / タイプ: ligand / ID: 4 / コピー数: 8 / 式: HOH |
|---|---|
| 分子量 | 理論値: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 QUANTUM (4k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




