+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of G6PT1 in complex with Glucose-6-phosphate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein structure / STRUCTURAL PROTEIN / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglucose-6-phosphate transmembrane transporter activity / Glycogen storage disease type Ib (SLC37A4) / glucose 6-phosphate:phosphate antiporter activity / glucose-6-phosphate transport / Gluconeogenesis / phosphate ion transmembrane transport / gluconeogenesis / glucose metabolic process / glucose homeostasis / endoplasmic reticulum membrane ...glucose-6-phosphate transmembrane transporter activity / Glycogen storage disease type Ib (SLC37A4) / glucose 6-phosphate:phosphate antiporter activity / glucose-6-phosphate transport / Gluconeogenesis / phosphate ion transmembrane transport / gluconeogenesis / glucose metabolic process / glucose homeostasis / endoplasmic reticulum membrane / endoplasmic reticulum / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Zhao Y / Chen Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Transport and inhibition mechanisms of G6PT1 Authors: Zhao Y / Chen Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63190.map.gz emd_63190.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63190-v30.xml emd-63190-v30.xml emd-63190.xml emd-63190.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63190.png emd_63190.png | 127 KB | ||
| Filedesc metadata |  emd-63190.cif.gz emd-63190.cif.gz | 5.5 KB | ||
| Others |  emd_63190_half_map_1.map.gz emd_63190_half_map_1.map.gz emd_63190_half_map_2.map.gz emd_63190_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63190 http://ftp.pdbj.org/pub/emdb/structures/EMD-63190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63190 | HTTPS FTP |
-Related structure data
| Related structure data |  9ll0MC  9ll1C  9lnbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63190.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63190.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_63190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of G6PT1 in complex with Glucose-6-phosphate
| Entire | Name: Cryo-EM structure of G6PT1 in complex with Glucose-6-phosphate |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of G6PT1 in complex with Glucose-6-phosphate
| Supramolecule | Name: Cryo-EM structure of G6PT1 in complex with Glucose-6-phosphate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glucose-6-phosphate exchanger SLC37A4
| Macromolecule | Name: Glucose-6-phosphate exchanger SLC37A4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.444695 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GYGYYRTVIF SAMFGGYSLY YFNRKTFSFV MPSLVEEIPL DKDDLGFITS SQSAAYAISK FVSGVLSDQM SARWLFSSGL LLVGLVNIF FAWSSTVPVF AALWFLNGLA QGLGWPPCGK VLRKWFEPSQ FGTWWAILST SMNLAGGLGP ILATILAQSY S WRSTLALS ...String: GYGYYRTVIF SAMFGGYSLY YFNRKTFSFV MPSLVEEIPL DKDDLGFITS SQSAAYAISK FVSGVLSDQM SARWLFSSGL LLVGLVNIF FAWSSTVPVF AALWFLNGLA QGLGWPPCGK VLRKWFEPSQ FGTWWAILST SMNLAGGLGP ILATILAQSY S WRSTLALS GALCVVVSFL CLLLIHNEPA DVGLRNLDPM PSEGKKGSLK EESTLQELLL SPYLWVLSTG YLVVFGVKTC CT DWGQFFL IQEKGQSALV GSSYMSALEV GGLVGSIAAG YLSDRAMAKA GLSNYGNPRH GLLLFMMAGM TVSMYLFRVT VTS DSPKLW ILVLGAVFGF SSYGPIALFG VIANESAPPN LCGTSHAIVG LMANVGGFLA GLPFSTIAKH YSWSTAFWVA EVIC AASTA AFFLLRNIRT KMGRV UniProtKB: Glucose-6-phosphate exchanger SLC37A4 |
-Macromolecule #2: 6-O-phosphono-beta-D-glucopyranose
| Macromolecule | Name: 6-O-phosphono-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 1 / Formula: BG6 |
|---|---|
| Molecular weight | Theoretical: 260.136 Da |
| Chemical component information | 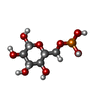 ChemComp-BG6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































