+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | bovine ABCC1 bound to estrogen sulfate and GSH | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / inward-open ABCC1 / estrogen sulfate / GSH / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationSphingolipid de novo biosynthesis / Heme degradation / Synthesis of Leukotrienes (LT) and Eoxins (EX) / Transport of RCbl within the body / cyclic nucleotide transport / Paracetamol ADME / ABC-family proteins mediated transport / glutathione transmembrane transport / leukotriene transport / ABC-type glutathione S-conjugate transporter activity ...Sphingolipid de novo biosynthesis / Heme degradation / Synthesis of Leukotrienes (LT) and Eoxins (EX) / Transport of RCbl within the body / cyclic nucleotide transport / Paracetamol ADME / ABC-family proteins mediated transport / glutathione transmembrane transport / leukotriene transport / ABC-type glutathione S-conjugate transporter activity / Cytoprotection by HMOX1 / ABC-type glutathione-S-conjugate transporter / glutathione transmembrane transporter activity / ABC-type xenobiotic transporter / ABC-type xenobiotic transporter activity / xenobiotic transport / lipid transport / xenobiotic transmembrane transporter activity / ABC-type transporter activity / positive regulation of inflammatory response / basolateral plasma membrane / response to xenobiotic stimulus / ATP hydrolysis activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Sun PP / Liu KX / Gao P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: bovine ABCC1 bound to estrogen sulfate and GSH Authors: Sun PP / Liu KX / Gao P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63057.map.gz emd_63057.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63057-v30.xml emd-63057-v30.xml emd-63057.xml emd-63057.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_63057_fsc.xml emd_63057_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_63057.png emd_63057.png | 40.3 KB | ||
| Filedesc metadata |  emd-63057.cif.gz emd-63057.cif.gz | 6.3 KB | ||
| Others |  emd_63057_half_map_1.map.gz emd_63057_half_map_1.map.gz emd_63057_half_map_2.map.gz emd_63057_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63057 http://ftp.pdbj.org/pub/emdb/structures/EMD-63057 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63057 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63057 | HTTPS FTP |
-Related structure data
| Related structure data |  9lg8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63057.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63057.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_63057_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_63057_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : inward-open bABCC1 bound to E1SO4 and GSH
| Entire | Name: inward-open bABCC1 bound to E1SO4 and GSH |
|---|---|
| Components |
|
-Supramolecule #1: inward-open bABCC1 bound to E1SO4 and GSH
| Supramolecule | Name: inward-open bABCC1 bound to E1SO4 and GSH / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Multidrug resistance-associated protein 1
| Macromolecule | Name: Multidrug resistance-associated protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 175.004688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALRDFCSVD GSDLFWEWNV TWNTSNPDFT KCFQNTVLVW VPCSYLWVCF PFYFLYLSHH DRGYIQMTHL NKAKTALGFL LWIVCWADL FYSFWERSMG KLLAPVFLVS PTLLGITMLL ATFLIQIERR RGVQSSGIML TFWLIALLCA LAILRSKIMT A LKEDARVD ...String: MALRDFCSVD GSDLFWEWNV TWNTSNPDFT KCFQNTVLVW VPCSYLWVCF PFYFLYLSHH DRGYIQMTHL NKAKTALGFL LWIVCWADL FYSFWERSMG KLLAPVFLVS PTLLGITMLL ATFLIQIERR RGVQSSGIML TFWLIALLCA LAILRSKIMT A LKEDARVD VFRDVTFYIY FSLVLIQLVL SCFSDRSPLF SETINDPNPC PESSASFLSR ITFWWITGMM VQGYRQPLES TD LWSLNKE DTSEQVVPVL VKNWKKECAK SRKQPVKIVY SSKDPAKPKG SSKVDVNEEA EALIVKCPQK ERDPSLFKVL YKT FGPYFL MSFLFKAVHD LMMFAGPEIL KLLINFVNDK KAPEWQGYFY TALLFISACL QTLVLHQYFH ICFVSGMRIK TAVI GAVYR KALVITNAAR KSSTVGEIVN LMSVDAQRFM DLATYINMIW SAPLQVILAL YLLWLNLGPS VLAGVAVMVL MVPLN AVMA MKTKTYQVAH MKSKDNRIKL MNEILNGIKV LKLYAWELAF KDKVLAIRQE ELKVLKKSAY LAAVGTFTWV CTPFLV ALS TFAVYVTVDE NNILDAQKAF VSLALFNILR FPLNILPMVI SSIVQASVSL KRLRVFLSHE DLDPDSIQRR PIKDAGA TN SITVKNATFT WARNDPPTLH GITFSVPEGS LVAVVGQVGC GKSSLLSALL AEMDKVEGHV TVKGSVAYVP QQAWIQNI S LRENILFGRQ LQERYYKAVV EACALLPDLE ILPSGDRTEI GEKGVNLSGG QKQRVSLARA VYCDSDVYLL DDPLSAVDA HVGKHIFENV IGPKGLLKNK TRLLVTHAIS YLPQMDVIIV MSGGKISEMG SYQELLARDG AFAEFLRTYA SAEQEQGQPE DGLAGVGGP GKEVKQMENG MLVTDTAGKQ MQRQLSSSSS YSRDVSQHHT STAELRKPGP TEETWKLVEA DKAQTGQVKL S VYWDYMKA IGLFISFLSI FLFLCNHVAS LVSNYWLSLW TDDPIVNGTQ EHTQVRLSVY GALGISQGIT VFGYSMAVSI GG IFASRRL HLDLLHNVLR SPISFFERTP SGNLVNRFSK ELDTVDSMIP QVIKMFMGSL FNVIGACIII LLATPMAAVI IPP LGLIYF FVQRFYVASS RQLKRLESVS RSPVYSHFNE TLLGVSVIRA FEEQERFIRQ SDLKVDENQK AYYPSIVANR WLAV RLECV GNCIVLFASL FAVISRHSLS AGLVGLSVSY SLQVTTYLNW LVRMSSEMET NIVAVERLKE YSETEKEAPW QIQDM APPK DWPQVGRVEF RDYGLRYRED LDLVLKHINV TIDGGEKVGI VGRTGAGKSS LTLGLFRIKE SAEGEIIIDD INIAKI GLH DLRFKITIIP QDPVLFSGSL RMNLDPFSQY SDEEVWTSLE LAHLKGFVSA LPDKLNHECA EGGENLSVGQ RQLVCLA RA LLRKTKILVL DEATAAVDLE TDDLIQSTIR TQFDDCTVLT IAHRLNTIMD YTRVIVLDKG EIQEWGSPSD LLQQRGLF Y SMAKDSGLVK LGSENLYFQG GSGGSGHHHH HHHHHHH UniProtKB: Multidrug resistance-associated protein 1 |
-Macromolecule #2: estrone 3-sulfate
| Macromolecule | Name: estrone 3-sulfate / type: ligand / ID: 2 / Number of copies: 1 / Formula: FY5 |
|---|---|
| Molecular weight | Theoretical: 350.429 Da |
| Chemical component information | 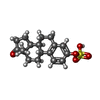 ChemComp-FY5: |
-Macromolecule #3: GLUTATHIONE
| Macromolecule | Name: GLUTATHIONE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GSH |
|---|---|
| Molecular weight | Theoretical: 307.323 Da |
| Chemical component information |  ChemComp-GSH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































