[English] 日本語
 Yorodumi
Yorodumi- EMDB-62884: Structure of beta1-AR-Gs complex bound to epinephrine and an allo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of beta1-AR-Gs complex bound to epinephrine and an allosteric modulator | |||||||||
 Map data Map data | Sharpened by DeepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationbeta-adrenergic receptor activity / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / beta1-adrenergic receptor activity / : / alpha-2A adrenergic receptor binding / regulation of circadian sleep/wake cycle, sleep / sensory perception of chemical stimulus / fear response / mu-type opioid receptor binding ...beta-adrenergic receptor activity / positive regulation of heart rate by epinephrine-norepinephrine / positive regulation of the force of heart contraction by epinephrine-norepinephrine / beta1-adrenergic receptor activity / : / alpha-2A adrenergic receptor binding / regulation of circadian sleep/wake cycle, sleep / sensory perception of chemical stimulus / fear response / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / heat generation / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Adrenoceptors / G alpha (q) signalling events / beta-2 adrenergic receptor binding / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / negative regulation of multicellular organism growth / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / diet induced thermogenesis / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / photoreceptor outer segment / D1 dopamine receptor binding / neuronal dense core vesicle / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / cardiac muscle cell apoptotic process / photoreceptor inner segment / ionotropic glutamate receptor binding / brown fat cell differentiation / response to cold / adenylate cyclase activator activity / PDZ domain binding / Schaffer collateral - CA1 synapse / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / positive regulation of cold-induced thermogenesis / signaling receptor complex adaptor activity / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / cell body / GTPase binding / cellular response to hypoxia / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / early endosome / cell population proliferation / positive regulation of MAPK cascade Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Sun Q / He G / Xu X / Liu X | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of beta1-AR-Gs complex bound to epinephrine and an allosteric modulator Authors: Sun Q / He G / Xu X / Liu X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62884.map.gz emd_62884.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62884-v30.xml emd-62884-v30.xml emd-62884.xml emd-62884.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_62884.png emd_62884.png | 23.6 KB | ||
| Filedesc metadata |  emd-62884.cif.gz emd-62884.cif.gz | 6.9 KB | ||
| Others |  emd_62884_additional_1.map.gz emd_62884_additional_1.map.gz emd_62884_half_map_1.map.gz emd_62884_half_map_1.map.gz emd_62884_half_map_2.map.gz emd_62884_half_map_2.map.gz | 57 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62884 http://ftp.pdbj.org/pub/emdb/structures/EMD-62884 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62884 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62884 | HTTPS FTP |
-Related structure data
| Related structure data |  9l84MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62884.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62884.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened by DeepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened by DeepEMhancer
| File | emd_62884_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Sharpened by DeepEMhancer
| File | emd_62884_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Sharpened by DeepEMhancer
| File | emd_62884_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of beta1-AR-Gs complex bound to epinephrine and an allo...
| Entire | Name: Structure of beta1-AR-Gs complex bound to epinephrine and an allosteric modulator |
|---|---|
| Components |
|
-Supramolecule #1: Structure of beta1-AR-Gs complex bound to epinephrine and an allo...
| Supramolecule | Name: Structure of beta1-AR-Gs complex bound to epinephrine and an allosteric modulator type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.769527 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQD DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGGQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ EALNLFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: NB35
| Macromolecule | Name: NB35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #5: Beta-1 adrenergic receptor
| Macromolecule | Name: Beta-1 adrenergic receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.275547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAGVLVLGA SEPGNLSSAA PLPDGAATAA RLLVPASPPA SLLPPASESP EPLSQQWTAG MGLLMALIVL LIVAGNVLVI VAIAKTPRL QTLTNLFIMS LASADLVMGL LVVPFGATIV VWGRWEYGSF FCELWTSVDV LCVTASIETL CVIALDRYLA I TSPFRYQS ...String: MGAGVLVLGA SEPGNLSSAA PLPDGAATAA RLLVPASPPA SLLPPASESP EPLSQQWTAG MGLLMALIVL LIVAGNVLVI VAIAKTPRL QTLTNLFIMS LASADLVMGL LVVPFGATIV VWGRWEYGSF FCELWTSVDV LCVTASIETL CVIALDRYLA I TSPFRYQS LLTRARARGL VCTVWAISAL VSFLPILMHW WRAESDEARR CYNDPKCCDF VTNRAYAIAS SVVSFYVPLC IM AFVYLRV FREAQKQVKK IDSCERRFLG GPARPPSPSP SPVPAPAPPP GPPRPAAAAA TAPLANGRAG KRRPSRLVAL REQ KALKTL GIIMGVFTLC WLPFFLANVV KAFHRELVPD RLFVFFNWLG YANSAFNPII YCRSPDFRKA FQGLLCCARR AARR RHATH GDRPRASGCL ARPGPPPSPG AASDDDDDDV VGATPPARLL EPWAGCNGGA AADSDSSLDE PCRPGFASES KV UniProtKB: Beta-1 adrenergic receptor |
-Macromolecule #6: (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9...
| Macromolecule | Name: (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER type: ligand / ID: 6 / Number of copies: 1 / Formula: DR7 |
|---|---|
| Molecular weight | Theoretical: 704.855 Da |
-Macromolecule #7: L-EPINEPHRINE
| Macromolecule | Name: L-EPINEPHRINE / type: ligand / ID: 7 / Number of copies: 1 / Formula: ALE |
|---|---|
| Molecular weight | Theoretical: 183.204 Da |
| Chemical component information | 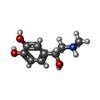 ChemComp-ALE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































