+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GPR119-Gs complex bound to AR231453 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / phosphatidylcholine binding / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding ...Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) / phosphatidylcholine binding / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / regulation of metabolic process / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / insulin secretion / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / photoreceptor outer segment / intracellular transport / D1 dopamine receptor binding / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / cardiac muscle cell apoptotic process / photoreceptor inner segment / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / G protein-coupled receptor activity / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Glucagon-type ligand receptors / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / positive regulation of cold-induced thermogenesis / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / cell body / retina development in camera-type eye / G protein activity / GTPase binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /     | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Sun Q / He G / Xu X / Liu X | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of GPR119-Gs complex bound to AR231453 Authors: Sun Q / He G / Xu X / Liu X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62871.map.gz emd_62871.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62871-v30.xml emd-62871-v30.xml emd-62871.xml emd-62871.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_62871.png emd_62871.png | 33.1 KB | ||
| Filedesc metadata |  emd-62871.cif.gz emd-62871.cif.gz | 6.7 KB | ||
| Others |  emd_62871_half_map_1.map.gz emd_62871_half_map_1.map.gz emd_62871_half_map_2.map.gz emd_62871_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62871 http://ftp.pdbj.org/pub/emdb/structures/EMD-62871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62871 | HTTPS FTP |
-Related structure data
| Related structure data |  9l79MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62871.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62871.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_62871_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_62871_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of GPR119-Gs complex bound to AR231453
| Entire | Name: Structure of GPR119-Gs complex bound to AR231453 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of GPR119-Gs complex bound to AR231453
| Supramolecule | Name: Structure of GPR119-Gs complex bound to AR231453 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #4, #2-#3, #6, #1, #5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glucose-dependent insulinotropic receptor
| Macromolecule | Name: Glucose-dependent insulinotropic receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.675809 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAMESSFS FGVILAVLAS LIIATNTLVA VAVLLLIHKN DGVSLCFTLN LAVADTLIGV AISGLLTDQ LSSPSRPTQK TLCSLRMAFV TSSAAASVLT VMLITFDRYL AIKQPFRYLK IMSGFVAGAC IAGLWLVSYL I GFLPLGIP ...String: MKTIIALSYI FCLVFADYKD DDDAMESSFS FGVILAVLAS LIIATNTLVA VAVLLLIHKN DGVSLCFTLN LAVADTLIGV AISGLLTDQ LSSPSRPTQK TLCSLRMAFV TSSAAASVLT VMLITFDRYL AIKQPFRYLK IMSGFVAGAC IAGLWLVSYL I GFLPLGIP MFQQTAYKGQ CSFFAVFHPH FVLTLSCVGF FPAMLLFVFF YCDMLKIASM HSQQIRKMEH AGAMAGGYRS PR TPSDFKA LRTVSVLIGS FALSWTPFLI TGIVQVACQE CHLYLVLERY LWLLGVGNSL LNPLIYAYWQ KEVRLQLYHM ALG VKKVLT SFLLFLSARN CGPERPRESS CHIVTISSSE FDG UniProtKB: Glucose-dependent insulinotropic receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  Trichoplusia (butterflies/moths) Trichoplusia (butterflies/moths) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.299104 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GCTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR ...String: GCTLSAEDKA AVERSKMIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIFE TKFQVDKVNF HMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNDFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI E DYFPEFAR YTTPEDATPE PGEDPRVTRA KYFIRDEFLR ISTASGDGRH YCYPHFTCAV DTENARRIFN DCRDIIQRMH LR QYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  Trichoplusia (butterflies/moths) Trichoplusia (butterflies/moths) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Macromolecule #6: NB35
| Macromolecule | Name: NB35 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.140742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHHEPEA |
-Macromolecule #7: N-(2-fluoranyl-4-methylsulfonyl-phenyl)-5-nitro-6-[4-(3-propan-2-...
| Macromolecule | Name: N-(2-fluoranyl-4-methylsulfonyl-phenyl)-5-nitro-6-[4-(3-propan-2-yl-1,2,4-oxadiazol-5-yl)piperidin-1-yl]pyrimidin-4-amine type: ligand / ID: 7 / Number of copies: 1 / Formula: 8WL |
|---|---|
| Molecular weight | Theoretical: 505.523 Da |
| Chemical component information | 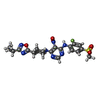 ChemComp-8WL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































