[English] 日本語
 Yorodumi
Yorodumi- EMDB-6224: CryoEM single particle reconstruction of anthrax toxin protective... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6224 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | |||||||||
 Map data Map data | Single particle reconstruction of intact anthrax toxin protective antigen pore | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial toxin / anthrax toxin / protective antigen / protein translocation channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host MAPK cascade / host cell cytosol / Uptake and function of anthrax toxins / host cell endosome membrane / protein homooligomerization / toxin activity / host cell plasma membrane / extracellular region / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Jiang J / Pentelute BL / Collier RJ / Zhou ZH | |||||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Authors: Jiansen Jiang / Bradley L Pentelute / R John Collier / Z Hong Zhou /  Abstract: Anthrax toxin, comprising protective antigen, lethal factor, and oedema factor, is the major virulence factor of Bacillus anthracis, an agent that causes high mortality in humans and animals. ...Anthrax toxin, comprising protective antigen, lethal factor, and oedema factor, is the major virulence factor of Bacillus anthracis, an agent that causes high mortality in humans and animals. Protective antigen forms oligomeric prepores that undergo conversion to membrane-spanning pores by endosomal acidification, and these pores translocate the enzymes lethal factor and oedema factor into the cytosol of target cells. Protective antigen is not only a vaccine component and therapeutic target for anthrax infections but also an excellent model system for understanding the mechanism of protein translocation. On the basis of biochemical and electrophysiological results, researchers have proposed that a phi (Φ)-clamp composed of phenylalanine (Phe)427 residues of protective antigen catalyses protein translocation via a charge-state-dependent Brownian ratchet. Although atomic structures of protective antigen prepores are available, how protective antigen senses low pH, converts to active pore, and translocates lethal factor and oedema factor are not well defined without an atomic model of its pore. Here, by cryo-electron microscopy with direct electron counting, we determine the protective antigen pore structure at 2.9-Å resolution. The structure reveals the long-sought-after catalytic Φ-clamp and the membrane-spanning translocation channel, and supports the Brownian ratchet model for protein translocation. Comparisons of four structures reveal conformational changes in prepore to pore conversion that support a multi-step mechanism by which low pH is sensed and the membrane-spanning channel is formed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
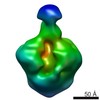
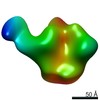
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6224.map.gz emd_6224.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6224-v30.xml emd-6224-v30.xml emd-6224.xml emd-6224.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6224.gif 400_6224.gif 80_6224.gif 80_6224.gif | 53.4 KB 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6224 http://ftp.pdbj.org/pub/emdb/structures/EMD-6224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6224 | HTTPS FTP |
-Validation report
| Summary document |  emd_6224_validation.pdf.gz emd_6224_validation.pdf.gz | 387.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6224_full_validation.pdf.gz emd_6224_full_validation.pdf.gz | 386.8 KB | Display | |
| Data in XML |  emd_6224_validation.xml.gz emd_6224_validation.xml.gz | 5.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6224 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6224 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6224 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6224 | HTTPS FTP |
-Related structure data
| Related structure data |  3j9cMC  6225C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6224.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6224.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single particle reconstruction of intact anthrax toxin protective antigen pore | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
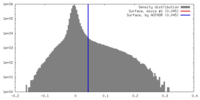
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Heptamer of C-terminal 63-kDa fragment of anthrax toxin protectiv...
| Entire | Name: Heptamer of C-terminal 63-kDa fragment of anthrax toxin protective antigen, pore conformation |
|---|---|
| Components |
|
-Supramolecule #1000: Heptamer of C-terminal 63-kDa fragment of anthrax toxin protectiv...
| Supramolecule | Name: Heptamer of C-terminal 63-kDa fragment of anthrax toxin protective antigen, pore conformation type: sample / ID: 1000 / Oligomeric state: Heptamer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 440 KDa |
-Macromolecule #1: Anthrax toxin protective antigen
| Macromolecule | Name: Anthrax toxin protective antigen / type: protein_or_peptide / ID: 1 / Name.synonym: PA / Number of copies: 7 / Oligomeric state: Heptamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 63 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Protective antigen |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 5 / Details: 50 mM NaOAc, 0.05% Igepal CA-630 |
| Grid | Details: Quantifoil R1.2/1.3 grid with thin carbon support, glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Jan 8, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 7062 / Average electron dose: 30 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 39062 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.1 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: OTHER / Software - Name: RELION / Number images used: 60455 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)