[English] 日本語
 Yorodumi
Yorodumi- EMDB-62150: Cryo-EM structure of human taurine transporter TAUT bound taurine... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human taurine transporter TAUT bound taurine in an occluded state in the saposin-disc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtaurine transmembrane transporter activity / alanine transmembrane transporter activity / gamma-aminobutyric acid transmembrane transporter activity / taurine:sodium symporter activity / import across plasma membrane / gamma-aminobutyric acid import / gamma-aminobutyric acid:sodium:chloride symporter activity / alanine transport / amino acid:sodium symporter activity / amino acid import across plasma membrane ...taurine transmembrane transporter activity / alanine transmembrane transporter activity / gamma-aminobutyric acid transmembrane transporter activity / taurine:sodium symporter activity / import across plasma membrane / gamma-aminobutyric acid import / gamma-aminobutyric acid:sodium:chloride symporter activity / alanine transport / amino acid:sodium symporter activity / amino acid import across plasma membrane / taurine transmembrane transport / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / SLC-mediated transport of neurotransmitters / neurotransmitter transport / microvillus membrane / amino acid transport / transport across blood-brain barrier / sodium ion transmembrane transport / cell projection / positive regulation of cell differentiation / modulation of chemical synaptic transmission / GABA-ergic synapse / basolateral plasma membrane / postsynaptic membrane / apical plasma membrane / neuronal cell body / dendrite / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang H / Xu EH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res. / Year: 2025 Journal: Cell Res. / Year: 2025Title: Cryo-EM structure of human taurine transporter TAUT bound taurine in an occluded state in the saposin-disc Authors: Zhang H / Xu EH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62150.map.gz emd_62150.map.gz | 85.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62150-v30.xml emd-62150-v30.xml emd-62150.xml emd-62150.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_62150_fsc.xml emd_62150_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_62150.png emd_62150.png | 86.2 KB | ||
| Filedesc metadata |  emd-62150.cif.gz emd-62150.cif.gz | 5.5 KB | ||
| Others |  emd_62150_half_map_1.map.gz emd_62150_half_map_1.map.gz emd_62150_half_map_2.map.gz emd_62150_half_map_2.map.gz | 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62150 http://ftp.pdbj.org/pub/emdb/structures/EMD-62150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62150 | HTTPS FTP |
-Related structure data
| Related structure data |  9k7nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62150.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62150.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_62150_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_62150_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sodium- and chloride-dependent taurine transporter
| Entire | Name: Sodium- and chloride-dependent taurine transporter |
|---|---|
| Components |
|
-Supramolecule #1: Sodium- and chloride-dependent taurine transporter
| Supramolecule | Name: Sodium- and chloride-dependent taurine transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium- and chloride-dependent taurine transporter
| Macromolecule | Name: Sodium- and chloride-dependent taurine transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.211 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKG SGMATKEKLQ CLKDFHKDIL KPSPGKSPGT RPEDEAEGKP PQREKWSSKI DFVLSVAGGF VGLGNVWRFP YLCYKNGGG AFLIPYFIFL FGSGLPVFFL EIIIGQYTSE GGITCWEKIC PLFSGIGYAS VVIVSLLNVY YIVILAWATY Y LFQSFQKE ...String: MDYKDDDDKG SGMATKEKLQ CLKDFHKDIL KPSPGKSPGT RPEDEAEGKP PQREKWSSKI DFVLSVAGGF VGLGNVWRFP YLCYKNGGG AFLIPYFIFL FGSGLPVFFL EIIIGQYTSE GGITCWEKIC PLFSGIGYAS VVIVSLLNVY YIVILAWATY Y LFQSFQKE LPWAHCNHSW NTPHCMEDTM RKNKSVWITI SSTNFTSPVI EFWERNVLSL SPGIDHPGSL KWDLALCLLL VW LVCFFCI WKGVRSTGKV VYFTATFPFA MLLVLLVRGL TLPGAGAGIK FYLYPDITRL EDPQVWIDAG TQIFFSYAIC LGA MTSLGS YNKYKYNSYR DCMLLGCLNS GTSFVSGFAI FSILGFMAQE QGVDIADVAE SGPGLAFIAY PKAVTMMPLP TFWS ILFFI MLLLLGLDSQ FVEVEGQITS LVDLYPSFLR KGYRREIFIA FVCSISYLLG LTMVTEGGMY VFQLFDYYAA SGVCL LWVA FFECFVIAWI YGGDNLYDGI EDMIGYRPGP WMKYSWAVIT PVLCVGCFIF SLVKYVPLTY NKTYVYPNWA IGLGWS LAL SSMLCVPLVI VIRLCQTEGP FLVRVKYLLT PREPNRWAVE REGATPYNSR TVMNGALVKP THIIVETMM UniProtKB: Sodium- and chloride-dependent taurine transporter |
-Macromolecule #2: 2-AMINOETHANESULFONIC ACID
| Macromolecule | Name: 2-AMINOETHANESULFONIC ACID / type: ligand / ID: 2 / Number of copies: 1 / Formula: TAU |
|---|---|
| Molecular weight | Theoretical: 125.147 Da |
| Chemical component information | 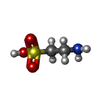 ChemComp-TAU: |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 50 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































