+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | core proteins of mature T7 | |||||||||||||||||||||
 Map data Map data | core proteins of T7 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont genome ejection through host cell envelope / : / symbiont entry into host cell via disruption of host cell wall peptidoglycan / peptidoglycan lytic transglycosylase activity / symbiont genome ejection through host cell envelope, short tail mechanism / peptidoglycan metabolic process / symbiont entry into host cell via disruption of host cell envelope / symbiont entry into host / T=7 icosahedral viral capsid / virion component ...symbiont genome ejection through host cell envelope / : / symbiont entry into host cell via disruption of host cell wall peptidoglycan / peptidoglycan lytic transglycosylase activity / symbiont genome ejection through host cell envelope, short tail mechanism / peptidoglycan metabolic process / symbiont entry into host cell via disruption of host cell envelope / symbiont entry into host / T=7 icosahedral viral capsid / virion component / killing of cells of another organism / defense response to bacterium / hydrolase activity / host cell plasma membrane / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |   Escherichia phage T7 (virus) Escherichia phage T7 (virus) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Liu HR / Chen WY | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2025 Journal: J Virol / Year: 2025Title: Conformational changes in and translocation of small proteins: insights into the ejection mechanism of podophages. Authors: Jing Zheng / Hao Xiao / Hao Pang / Li Wang / Jingdong Song / Wenyuan Chen / Lingpeng Cheng / Hongrong Liu /  Abstract: Podophage tails are too short to span the cell envelope during infection. Consequently, podophages initially eject the core proteins within the head for the formation of an elongated trans-envelope ...Podophage tails are too short to span the cell envelope during infection. Consequently, podophages initially eject the core proteins within the head for the formation of an elongated trans-envelope channel for DNA ejection. Although the core proteins of bacteriophage T7 have been resolved at near-atomic resolution, the mechanisms of core proteins and DNA ejection remain to be fully elucidated. In this study, we provided improved structures of core proteins in mature T7 and the portal-tail complex in lipopolysaccharide-induced DNA-ejected T7 to resolutions of approximately 3 Å. Using these structures, we identified three small proteins, namely gp14, gp6.7, and gp7.3, and illustrated the conformational changes in and translocation of these proteins from the mature to DNA-ejected states. Our structures indicate that gp6.7, which participates in the assembly of the core and trans-envelope channel, is a core protein, and that gp7.3 serves as a structural scaffold to assist the assembly of the nozzle into the adaptor. IMPORTANCE: Podophage T7 core proteins form an elongated trans-envelope channel for genomic DNA delivery into the host cell. The structures of the core proteins within the mature T7 and assembled in ...IMPORTANCE: Podophage T7 core proteins form an elongated trans-envelope channel for genomic DNA delivery into the host cell. The structures of the core proteins within the mature T7 and assembled in the periplasmic tunnel form in the DNA-ejected T7 have been resolved previously. Here, we resolved the structures of two new structural proteins (gp6.7 and gp7.3) within mature T7 and receptor-induced DNA-ejected T7. The gp6.7 protein participates in the assembly of the core complex within mature T7 and the trans-envelope channel during T7 infection; therefore, gp6.7 is a core protein. Before T7 infection, gp7.3 plays a role in promoting the assembly of the nozzle into the adaptor. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61909.map.gz emd_61909.map.gz | 115.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61909-v30.xml emd-61909-v30.xml emd-61909.xml emd-61909.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61909.png emd_61909.png | 164.5 KB | ||
| Filedesc metadata |  emd-61909.cif.gz emd-61909.cif.gz | 7 KB | ||
| Others |  emd_61909_half_map_1.map.gz emd_61909_half_map_1.map.gz emd_61909_half_map_2.map.gz emd_61909_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61909 http://ftp.pdbj.org/pub/emdb/structures/EMD-61909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61909 | HTTPS FTP |
-Related structure data
| Related structure data |  9jyyMC  9jyzC  9jz0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61909.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61909.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | core proteins of T7 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
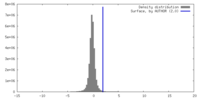
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_61909_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
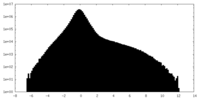
| Density Histograms |
-Half map: #1
| File | emd_61909_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
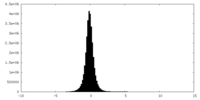
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage T7
| Entire | Name:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage T7
| Supramolecule | Name: Escherichia phage T7 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10760 / Sci species name: Escherichia phage T7 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Protein 6.7
| Macromolecule | Name: Protein 6.7 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 9.354512 KDa |
| Sequence | String: MCFSPKIKTP KMDTNQIRAV EPAPLTQEVS SVEFGGSSDE TDTEGTEVSG RKGLKVERDD SVAKSKASGN GSARMKSSIR KSAFGGKK UniProtKB: Protein 6.7 |
-Macromolecule #2: Internal virion protein gp14
| Macromolecule | Name: Internal virion protein gp14 / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 20.990842 KDa |
| Sequence | String: MCWAAAIPIA ISGAQAISGQ NAQAKMIAAQ TAAGRRQAME IMRQTNIQNA DLSLQARSKL EEASAELTSQ NMQKVQAIGS IRAAIGESM LEGSSMDRIK RVTEGQFIRE ANMVTENYRR DYQAIFAQQL GGTQSAASQI DEIYKSEQKQ KSKLQMVLDP L AIMGSSAA ...String: MCWAAAIPIA ISGAQAISGQ NAQAKMIAAQ TAAGRRQAME IMRQTNIQNA DLSLQARSKL EEASAELTSQ NMQKVQAIGS IRAAIGESM LEGSSMDRIK RVTEGQFIRE ANMVTENYRR DYQAIFAQQL GGTQSAASQI DEIYKSEQKQ KSKLQMVLDP L AIMGSSAA SAYASGAFDS KSTTKAPIVA AKGTKTGR UniProtKB: Internal virion protein gp14 |
-Macromolecule #3: Internal virion protein gp15
| Macromolecule | Name: Internal virion protein gp15 / type: protein_or_peptide / ID: 3 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 84.454008 KDa |
| Sequence | String: MSKIESALQA AQPGLSRLRG GAGGMGYRAA TTQAEQPRSS LLDTIGRFAK AGADMYTAKE QRARDLADER SNEIIRKLTP EQRREALNN GTLLYQDDPY AMEALRVKTG RNAAYLVDDD VMQKIKEGVF RTREEMEEYR HSRLQEGAKV YAEQFGIDPE D VDYQRGFN ...String: MSKIESALQA AQPGLSRLRG GAGGMGYRAA TTQAEQPRSS LLDTIGRFAK AGADMYTAKE QRARDLADER SNEIIRKLTP EQRREALNN GTLLYQDDPY AMEALRVKTG RNAAYLVDDD VMQKIKEGVF RTREEMEEYR HSRLQEGAKV YAEQFGIDPE D VDYQRGFN GDITERNISL YGAHDNFLSQ QAQKGAIMNS RVELNGVLQD PDMLRRPDSA DFFEKYIDNG LVTGAIPSDA QA TQLISQA FSDASSRAGG ADFLMRVGDK KVTLNGATTT YRELIGEEQW NALMVTAQRS QFETDAKLNE QYRLKINSAL NQE DPRTAW EMLQGIKAEL DKVQPDEQMT PQREWLISAQ EQVQNQMNAW TKAQAKALDD SMKSMNKLDV IDKQFQKRIN GEWV STDFK DMPVNENTGE FKHSDMVNYA NKKLAEIDSM DIPDGAKDAM KLKYLQADSK DGAFRTAIGT MVTDAGQEWS AAVIN GKLP ERTPAMDALR RIRNADPQLI AALYPDQAEL FLTMDMMDKQ GIDPQVILDA DRLTVKRSKE QRFEDDKAFE SALNAS KAP EIARMPASLR ESARKIYDSV KYRSGNESMA MEQMTKFLKE STYTFTGDDV DGDTVGVIPK NMMQVNSDPK SWEQGRD IL EEARKGIIAS NPWITNKQLT MYSQGDSIYL MDTTGQVRVR YDKELLSKVW SENQKKLEEK AREKALADVN KRAPIVAA T KAREAAAKRV REKRKQTPKF IYGRKE |
-Macromolecule #4: Peptidoglycan transglycosylase gp16
| Macromolecule | Name: Peptidoglycan transglycosylase gp16 / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO EC number: Lyases; Carbon-oxygen lyases; Acting on polysaccharides |
|---|---|
| Source (natural) | Organism:   Escherichia phage T7 (virus) Escherichia phage T7 (virus) |
| Molecular weight | Theoretical: 144.028219 KDa |
| Sequence | String: MDKYDKNVPS DYDGLFQKAA DANGVSYDLL RKVAWTESRF VPTAKSKTGP LGMMQFTKAT AKALGLRVTD GPDDDRLNPE LAINAAAKQ LAGLVGKFDG DELKAALAYN QGEGRLGNPQ LEAYSKGDFA SISEEGRNYM RNLLDVAKSP MAGQLETFGG I TPKGKGIP ...String: MDKYDKNVPS DYDGLFQKAA DANGVSYDLL RKVAWTESRF VPTAKSKTGP LGMMQFTKAT AKALGLRVTD GPDDDRLNPE LAINAAAKQ LAGLVGKFDG DELKAALAYN QGEGRLGNPQ LEAYSKGDFA SISEEGRNYM RNLLDVAKSP MAGQLETFGG I TPKGKGIP AEVGLAGIGH KQKVTQELPE STSFDVKGIE QEATAKPFAK DFWETHGETL DEYNSRSTFF GFKNAAEAEL SN SVAGMAF RAGRLDNGFD VFKDTITPTR WNSHIWTPEE LEKIRTEVKN PAYINVVTGG SPENLDDLIK LANENFENDS RAA EAGLGA KLSAGIIGAG VDPLSYVPMV GVTGKGFKLI NKALVVGAES AALNVASEGL RTSVAGGDAD YAGAALGGFV FGAG MSAIS DAVAAGLKRS KPEAEFDNEF IGPMMRLEAR ETARNANSAD LSRMNTENMK FEGEHNGVPY EDLPTERGAV VLHDG SVLS ASNPINPKTL KEFSEVDPEK AARGIKLAGF TEIGLKTLGS DDADIRRVAI DLVRSPTGMQ SGASGKFGAT ASDIHE RLH GTDQRTYNDL YKAMSDAMKD PEFSTGGAKM SREETRYTIY RRAALAIERP ELQKALTPSE RIVMDIIKRH FDTKREL ME NPAIFGNTKA VSIFPESRHK GTYVPHVYDR HAKALMIQRY GAEGLQEGIA RSWMNSYVSR PEVKARVDEM LKELHGVK E VTPEMVEKYA MDKAYGISHS DQFTNSSIIE ENIEGLVGIE NNSFLEARNL FDSDLSITMP DGQQFSVNDL RDFDMFRIM PAYDRRVNGD IAIMGSTGKT TKELKDEILA LKAKAEGDGK KTGEVHALMD TVKILTGRAR RNQDTVWETS LRAINDLGFF AKNAYMGAQ NITEIAGMIV TGNVRALGHG IPILRDTLYK SKPVSAKELK ELHASLFGKE VDQLIRPKRA DIVQRLREAT D TGPAVANI VGTLKYSTQE LAARSPWTKL LNGTTNYLLD AARQGMLGDV ISATLTGKTT RWEKEGFLRG ASVTPEQMAG IK SLIKEHM VRGEDGKFTV KDKQAFSMDP RAMDLWRLAD KVADEAMLRP HKVSLQDSHA FGALGKMVMQ FKSFTIKSLN SKF LRTFYD GYKNNRAIDA ALSIITSMGL AGGFYAMAAH VKAYALPKEK RKEYLERALD PTMIAHAALS RSSQLGAPLA MVDL VGGVL GFESSKMARS TILPKDTVKE RDPNKPYTSR EVMGAMGSNL LEQMPSAGFV ANVGATLMNA AGVVNSPNKA TEQDF MTGL MNSTKELVPN DPLTQQLVLK IYEANGVNLR ERRK UniProtKB: Peptidoglycan transglycosylase gp16 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)