+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CGP54626-bound-DGABAB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COMPLEX / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of synaptic vesicle fusion to presynaptic active zone membrane / olfactory behavior / G protein-coupled GABA receptor activity / cellular response to mechanical stimulus / negative regulation of insulin secretion / postsynaptic membrane / G protein-coupled receptor signaling pathway / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | shen CS / liu JF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis of GABAB receptor activation during evolution Authors: shen CS / liu JF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61742.map.gz emd_61742.map.gz | 78.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61742-v30.xml emd-61742-v30.xml emd-61742.xml emd-61742.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61742_fsc.xml emd_61742_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_61742.png emd_61742.png | 30.3 KB | ||
| Masks |  emd_61742_msk_1.map emd_61742_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61742.cif.gz emd-61742.cif.gz | 6.5 KB | ||
| Others |  emd_61742_half_map_1.map.gz emd_61742_half_map_1.map.gz emd_61742_half_map_2.map.gz emd_61742_half_map_2.map.gz | 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61742 http://ftp.pdbj.org/pub/emdb/structures/EMD-61742 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61742 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61742 | HTTPS FTP |
-Related structure data
| Related structure data |  9jqxMC  9jr0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61742.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61742.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61742_msk_1.map emd_61742_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61742_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61742_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Drosophila metabotropic GABA-B receptor
| Entire | Name: Drosophila metabotropic GABA-B receptor |
|---|---|
| Components |
|
-Supramolecule #1: Drosophila metabotropic GABA-B receptor
| Supramolecule | Name: Drosophila metabotropic GABA-B receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 kDa/nm |
-Macromolecule #1: Metabotropic GABA-B receptor subtype 1
| Macromolecule | Name: Metabotropic GABA-B receptor subtype 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 79.778414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ELHIGGIFPI AGKGGWQGGQ ACMPATRLAL DDVNKQPNLL PGFKLILHSN DSECEPGLGA SVMYNLLYNK PQKLMLLAGC STVCTTVAE AAKMWNLIVL CYGASSPALS DRKRFPTLFR THPSATVHNP TRIKLMKKFG WSRVAILQQA EEVFISTVED L ENRCMEAG ...String: ELHIGGIFPI AGKGGWQGGQ ACMPATRLAL DDVNKQPNLL PGFKLILHSN DSECEPGLGA SVMYNLLYNK PQKLMLLAGC STVCTTVAE AAKMWNLIVL CYGASSPALS DRKRFPTLFR THPSATVHNP TRIKLMKKFG WSRVAILQQA EEVFISTVED L ENRCMEAG VEIVTRQSFL SDPTDAVRNL RRQDARIIVG LFYVVAARRV LCEMYKQQLY GRAHVWFFIG WYEDNWYEVN LK AEGITCT VEQMRIAAEG HLTTEALMWN QNNQTTISGM TAEEFRHRLN QALIEEGYDI NHDRYPEGYQ EAPLAYDAVW SVA LAFNKT MERLTTGKKS LRDFTYTDKE IADEIYAAMN STQFLGVSGV VAFSSQGDRI ALTQIEQMID GKYEKLGYYD TQLD NLSWL NTEQWIGGKV PQDRTIVTHV LRTVSLPLFV CMCTISSCGI FVAFALIIFN IWNKHRRVIQ SSHPVCNTIM LFGVI ICLI SVILLGIDGR FVSPEEYPKI CQARAWLLST GFTLAYGAMF SKVWRVHRFT TKAKTDPKKK VEPWKLYTMV SGLLSI DLV ILLSWQIFDP LQRYLETFPL EDPVSTTDDI KIRPELEHCE SQRNSMWLGL VYGFKGLILV FGLFLAYETR SIKVKQI ND SRYVGMSIYN VVVLCLITAP VGMVIASQQD ASFAFVALAV IFCCFLSMLL IFVPKVIEVI RH UniProtKB: Metabotropic GABA-B receptor subtype 1 |
-Macromolecule #2: Gamma-aminobutyric acid type B receptor subunit 2
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.493703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RTAKRSDVYI AGFFPYGDGV ENSYTGRGVM PSVKLALGHV NEHGKILANY RLHMWWNDTQ CNAAVGVKSF FDMMHSGPNK VMLFGAACT HVTDPIAKAS KHWHLTQLSY ADTHPMFTKD AFPNFFRVVP SENAFNAPRL ALLKEFNWTR VGTVYQNEPR Y SLPHNHMV ...String: RTAKRSDVYI AGFFPYGDGV ENSYTGRGVM PSVKLALGHV NEHGKILANY RLHMWWNDTQ CNAAVGVKSF FDMMHSGPNK VMLFGAACT HVTDPIAKAS KHWHLTQLSY ADTHPMFTKD AFPNFFRVVP SENAFNAPRL ALLKEFNWTR VGTVYQNEPR Y SLPHNHMV ADLDAMEVEV VETQSFVNDV AESLKKLREK DVRIILGNFN EHFARKAFCE AYKLDMYGRA YQWLIMATYS TD WWNVTQD SECSVEEIAT ALEGAILVDL LPLSTSGDIT VAGITADEYL VEYDRLRGTE YSRFHGYTYD GIWAAALAIQ YVA EKREDL LTHFDYRVKD WESVFLEALR NTSFEGVTGP VRFYNNERKA NILINQFQLG QMEKIGEYHS QKSHLDLSLG KPVK WVGKT PPKDRTLIYI EHSQVNPTIY IVSASASVIG VIIATVFLAF NIKYRNQRYI KMSSPHLNNL IIVGCMITYL SIIFL GLDT TLSSVAAFPY ICTARAWILM AGFSLSFGAM FSKTWRVHSI FTDLKLNKKV IKDYQLFMVV GVLLAIDIAI ITTWQI ADP FYRETKQLEP LHHENIDDVL VIPENEYCQS EHMTIFVSII YAYKGLLLVF GAFLAWETRH VSIPALNDSK HIGFSVY NV FITCLAGAAI SLVLSDRKDL VFVLLSFFII FCTTATLCLV FVPKLVELKR NPQGVVDKRV RATLRPMSKN GRRDSSVC E LEQRLRDVKN TNCRFRKALM EKENELQALI RKLGPEARKW IDGVTC UniProtKB: Gamma-aminobutyric acid type B receptor subunit 2 |
-Macromolecule #3: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]...
| Macromolecule | Name: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid type: ligand / ID: 3 / Number of copies: 1 / Formula: 2BV |
|---|---|
| Molecular weight | Theoretical: 408.3 Da |
| Chemical component information | 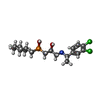 ChemComp-2BV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 7.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9jqx: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































