+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the METH-bound hTAAR1-Gs complex | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | cryo-EM / GPCR / Trace amine-associated receptor 1 / SIGNALING PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationAmine ligand-binding receptors / trace-amine receptor activity / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity ...Amine ligand-binding receptors / trace-amine receptor activity / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / endomembrane system / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / electron transport chain / bone development / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / endoplasmic reticulum membrane / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | ||||||||||||||||||||||||
 Authors Authors | Lin Y / Wang J / Shi F | ||||||||||||||||||||||||
| Funding support |  China, 7 items China, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2024 Journal: J Med Chem / Year: 2024Title: Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation. Authors: Yun Lin / Jiening Wang / Fan Shi / Linlin Yang / Shan Wu / Anna Qiao / Sheng Ye /  Abstract: Trace amine-associated receptor 1 (TAAR1), a member of the trace amine receptor family, recognizes various trace amines in the brain, including endogenous β-phenylethylamine (PEA) and ...Trace amine-associated receptor 1 (TAAR1), a member of the trace amine receptor family, recognizes various trace amines in the brain, including endogenous β-phenylethylamine (PEA) and methamphetamine (METH). TAAR1 is a novel target for several neurological disorders, including schizophrenia, depression, and substance abuse. Herein, we report the structure of the human TAAR1-G protein complex bound to METH. Using functional studies, we reveal the molecular basis of METH recognition by TAAR1, and potential mechanisms underlying the selectivity of TAAR1 for different ligands. Molecular dynamics simulations further elucidated possible mechanisms for the binding of chiral amphetamine (AMPH)-like psychoactive drugs to TAAR1. Additionally, we discovered a hydrophobic core on the transmembrane helices (TM), TM5 and TM6, explaining the unique mechanism of TAAR1 activation. These findings reveal the ligand recognition pattern and activation mechanism of TAAR1, which has important implications for the development of next-generation treatments for substance abuse and various neurological disorders. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61567.map.gz emd_61567.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61567-v30.xml emd-61567-v30.xml emd-61567.xml emd-61567.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61567_fsc.xml emd_61567_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_61567.png emd_61567.png | 36.8 KB | ||
| Filedesc metadata |  emd-61567.cif.gz emd-61567.cif.gz | 7.1 KB | ||
| Others |  emd_61567_half_map_1.map.gz emd_61567_half_map_1.map.gz emd_61567_half_map_2.map.gz emd_61567_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61567 http://ftp.pdbj.org/pub/emdb/structures/EMD-61567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61567 | HTTPS FTP |
-Validation report
| Summary document |  emd_61567_validation.pdf.gz emd_61567_validation.pdf.gz | 774.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_61567_full_validation.pdf.gz emd_61567_full_validation.pdf.gz | 774.2 KB | Display | |
| Data in XML |  emd_61567_validation.xml.gz emd_61567_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_61567_validation.cif.gz emd_61567_validation.cif.gz | 24.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61567 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61567 | HTTPS FTP |
-Related structure data
| Related structure data |  9jkqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61567.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61567.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.851 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61567_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61567_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : METH-bound hTAAR1-Gs complex
| Entire | Name: METH-bound hTAAR1-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: METH-bound hTAAR1-Gs complex
| Supramolecule | Name: METH-bound hTAAR1-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.540746 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGMKYLLPTA AAGLLLLAAQ PAMAQVQLQE SGGGLVQPGG SLRLSCAASG FTFSNYKMNW VRQAPGKGLE WVSDISQSGA SISYTGSVK GRFTISRDNA KNTLYLQMNS LKPEDTAVYY CARCPAPFTR DCFDVTSTTY AYRGQGTQVT VSSHHHHHHE P EA |
-Macromolecule #5: Soluble cytochrome b562,Trace amine-associated receptor 1
| Macromolecule | Name: Soluble cytochrome b562,Trace amine-associated receptor 1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.969645 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFAGAPA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLEDK SPDSPEMKDF RHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLMMPF CHNIINISCV KNNWSNDVRA SLYSLMVLII L TTLVGNLI ...String: MKTIIALSYI FCLVFAGAPA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLEDK SPDSPEMKDF RHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLMMPF CHNIINISCV KNNWSNDVRA SLYSLMVLII L TTLVGNLI VIVSISHFKQ LHTPTNWLIV SMATVDFLLG CLVMPYSMVR SAEHCWYFGE VFCKIHTSTD IMLSSASIWH LS FISIDRY YAVCDPLRYK AKMNILVICV MIFISWSVPA VFAFGMIFLE LNFKGAEEIY YKHVHCRGGC SVFFSKISGV LTF MTSFYI PGSIMLCVYY RIYLIAKEQA RLISDANQKL QIGLEMKNGI SQSKERKAVK TLGIVMGVFL ICWCPFFICT VMDP FLHYI IPPTLNDVLI WFGYLNSTFN PMVYAFFYPW FRKALKMMLF GKIFQKDSSR CKLFLELSSE FLEVLFQGPW SHPQF EKGG GSGGGSGGSA WSHPQFEKDY KDDDDK UniProtKB: Soluble cytochrome b562, Trace amine-associated receptor 1 |
-Macromolecule #6: (2S)-N-methyl-1-phenylpropan-2-amine
| Macromolecule | Name: (2S)-N-methyl-1-phenylpropan-2-amine / type: ligand / ID: 6 / Number of copies: 1 / Formula: B40 |
|---|---|
| Molecular weight | Theoretical: 149.233 Da |
| Chemical component information | 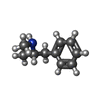 ChemComp-B40: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































