[English] 日本語
 Yorodumi
Yorodumi- EMDB-61504: Hepatitis E virus capsid protein E2s domain (genotype I) in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
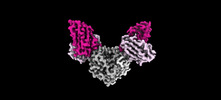
| Title | Hepatitis E virus capsid protein E2s domain (genotype I) in complex with Fab C6 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hepatitis E virus / Capsid protein / E2s domain / Immune complex / C6 antibody / VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum / T=1 icosahedral viral capsid / host cell surface / host cell Golgi apparatus / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell / host cell nucleus / structural molecule activity / RNA binding / extracellular region Similarity search - Function | |||||||||
| Biological species |  Paslahepevirus balayani / Paslahepevirus balayani /  Homo sapiens (human) / Homo sapiens (human) /  Hepatitis E virus genotype 1 (isolate Human/Burma) Hepatitis E virus genotype 1 (isolate Human/Burma) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Liu L / Zheng Q / Li S / Xia N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Hepatol / Year: 2025 Journal: J Hepatol / Year: 2025Title: Antibodies elicited by hepatitis E vaccination in humans confer cross-genus protection against rat hepatitis E virus. Authors: Zihao Chen / Liqin Liu / Jianwen Situ / Guanghui Li / Shaoqi Guo / Qi Lai / Shusheng Wu / Yanan Jiang / Jingyan Fan / Zimin Tang / Yu Li / Guiping Wen / Siling Wang / Dong Ying / Yonghao ...Authors: Zihao Chen / Liqin Liu / Jianwen Situ / Guanghui Li / Shaoqi Guo / Qi Lai / Shusheng Wu / Yanan Jiang / Jingyan Fan / Zimin Tang / Yu Li / Guiping Wen / Siling Wang / Dong Ying / Yonghao Liang / Stanley Siu-Fung Ho / Xiaodan Ma / James Yiu-Hung Tsoi / Estie Hon-Kiu Shun / Nicholas Foo-Siong Chew / Weihui Ma / Weiwei Mao / Tingting Li / Zhenqin Chen / Mujin Fang / Yingbin Wang / Hai Yu / Fa Zhang / Anna Jinxia Zhang / Shaowei Li / Ningshao Xia / Siddharth Sridhar / Qingbing Zheng / Zizheng Zheng /  Abstract: BACKGROUND & AIMS: Paslahepevirus balayani (bHEV), also known as hepatitis E virus (HEV), encompasses eight genotypes, five of which infect humans. Rats are natural reservoirs of Rocahepevirus ratti ...BACKGROUND & AIMS: Paslahepevirus balayani (bHEV), also known as hepatitis E virus (HEV), encompasses eight genotypes, five of which infect humans. Rats are natural reservoirs of Rocahepevirus ratti genotype 1 (HEV-r-1; rat HEV; rHEV), which has recently been implicated in viral hepatitis. Despite the antigenic divergence between bHEV and rHEV, studies on shared protective antibodies remain rare. METHODS: Polyclonal and monoclonal antibody responses against bHEV and rHEV were analyzed using antibody enzyme-linked immunosorbent assays. The efficacy of six potent bHEV-elicited cross-reactive ...METHODS: Polyclonal and monoclonal antibody responses against bHEV and rHEV were analyzed using antibody enzyme-linked immunosorbent assays. The efficacy of six potent bHEV-elicited cross-reactive antibodies in preventing rHEV infection was evaluated via challenge assays in rats. Cryo-electron microscopy was performed to assess the structural basis for the differential protective efficacy of the six antibodies. The viral lysis ability of these antibodies was assessed by separately reacting purified HEV-b-1 and HEV-r-1 virions with each antibody. RESULTS: We determined that antibody responses to bHEV infection and vaccination possess limited cross-reactivity to rHEV and identified two cross-reactive antigenic sites within the E2s domain. ...RESULTS: We determined that antibody responses to bHEV infection and vaccination possess limited cross-reactivity to rHEV and identified two cross-reactive antigenic sites within the E2s domain. Structural analysis and animal challenge studies pinpointed potent cross-reactive antibodies targeting antigenic site 1, indicating its prophylactic efficacy against rHEV. Conversely, antibodies recognizing antigenic site 2 were found to facilitate viral lysis of bHEV but not rHEV. CONCLUSIONS: These findings underscore the importance of antigenic site 1 in the design of broad-spectrum vaccines and therapeutics to mitigate the impact of diverse HEV genotypes on human health. IMPACT AND IMPLICATIONS: Rat HEV (rHEV) spillover to humans represents an unprecedented threat. Significant antigenic differences between rHEV and bHEV (Paslahepevirus balayani) may exacerbate the ...IMPACT AND IMPLICATIONS: Rat HEV (rHEV) spillover to humans represents an unprecedented threat. Significant antigenic differences between rHEV and bHEV (Paslahepevirus balayani) may exacerbate the impact of viral hepatitis. Our study reveals that cross-reactive human antibodies can offer protection against rHEV infection. Cross-protective antibodies targeting antigenic site 1 can be used to guide the development of practical strategies for preventing rHEV infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61504.map.gz emd_61504.map.gz | 202 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61504-v30.xml emd-61504-v30.xml emd-61504.xml emd-61504.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61504.png emd_61504.png | 28 KB | ||
| Filedesc metadata |  emd-61504.cif.gz emd-61504.cif.gz | 5.9 KB | ||
| Others |  emd_61504_half_map_1.map.gz emd_61504_half_map_1.map.gz emd_61504_half_map_2.map.gz emd_61504_half_map_2.map.gz | 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61504 http://ftp.pdbj.org/pub/emdb/structures/EMD-61504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61504 | HTTPS FTP |
-Related structure data
| Related structure data |  9jifMC  9jieC  9jigC  9jiiC  9jijC  9jikC  9jilC  9jimC  9jinC  9jioC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61504.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61504.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
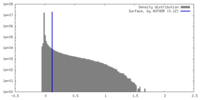
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_61504_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
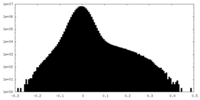
| Density Histograms |
-Half map: #1
| File | emd_61504_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HEV capsid protein E2s domain in complex with fab C6
| Entire | Name: HEV capsid protein E2s domain in complex with fab C6 |
|---|---|
| Components |
|
-Supramolecule #1: HEV capsid protein E2s domain in complex with fab C6
| Supramolecule | Name: HEV capsid protein E2s domain in complex with fab C6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Paslahepevirus balayani Paslahepevirus balayani |
-Supramolecule #2: HEV E2s domain
| Supramolecule | Name: HEV E2s domain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Paslahepevirus balayani Paslahepevirus balayani |
-Supramolecule #3: C6 Fab
| Supramolecule | Name: C6 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Secreted protein ORF2
| Macromolecule | Name: Secreted protein ORF2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hepatitis E virus genotype 1 (isolate Human/Burma) Hepatitis E virus genotype 1 (isolate Human/Burma) |
| Molecular weight | Theoretical: 22.956402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLFYSRPVVS ANGEPTVKLY TSVENAQQDK GIAIPHDIDL GESRVVIQDY DNQHEQDRPT PSPAPSRPFS VLRANDVLWL SLTAAEYDQ STYGSSTGPV YVSDSVTLVN VATGAQAVAR SLDWTKVTLD GRPLSTIQQY SKTFFVLPLR GKLSFWEAGT T KAGYPYNY ...String: QLFYSRPVVS ANGEPTVKLY TSVENAQQDK GIAIPHDIDL GESRVVIQDY DNQHEQDRPT PSPAPSRPFS VLRANDVLWL SLTAAEYDQ STYGSSTGPV YVSDSVTLVN VATGAQAVAR SLDWTKVTLD GRPLSTIQQY SKTFFVLPLR GKLSFWEAGT T KAGYPYNY NTTASDQLLV ENAAGHRVAI STYTTSLGAG PVSISAVAVL APHSA UniProtKB: Pro-secreted protein ORF2 |
-Macromolecule #2: C6 Fab heavy chain
| Macromolecule | Name: C6 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.60716 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFR SYAIHWVRQA PGKGLEWVAL ISYDGSNGYY ADSVKGRFTI SRDNSKNTVY LQVNTLRAE DTALYYCARD RGSIVEPAAL YIDYWGQGTL VTVSS |
-Macromolecule #3: C6 Fab light chain
| Macromolecule | Name: C6 Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.425632 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SAAPGQMVTI SCSGSSSNIG NNYVSWYQHL PGTAPKLLIY DNNKRPSGIP DRFSGSKSGT SVTLGITGLQ TGDEADYYC GTWDSSLSAV VFGGGTKLTV L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)