+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Human Kcnk13. | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | potassium channel / microglia function / K2P family / neurodegenerative diseases. / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of excitatory synapse pruning / Tandem pore domain halothane-inhibited K+ channel (THIK) / regulation of NLRP3 inflammasome complex assembly / Phase 4 - resting membrane potential / potassium ion leak channel activity / regulation of resting membrane potential / outward rectifier potassium channel activity / monoatomic ion channel complex / potassium channel activity / potassium ion transmembrane transport ...regulation of excitatory synapse pruning / Tandem pore domain halothane-inhibited K+ channel (THIK) / regulation of NLRP3 inflammasome complex assembly / Phase 4 - resting membrane potential / potassium ion leak channel activity / regulation of resting membrane potential / outward rectifier potassium channel activity / monoatomic ion channel complex / potassium channel activity / potassium ion transmembrane transport / protein heterodimerization activity / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||||||||
 Authors Authors | Baobin L / Ran Z / Jin W | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Gating mechanism of the two-pore-domain potassium channel THIK1. Authors: Xiangyun Fang / Haichao Jin / Jin Wang / Ran Zhang / Baobin Li /  Abstract: TWIK-related halothane-inhibited potassium channel (THIK1) maintains the resting membrane potential and regulates potassium efflux in microglia. It is a potential therapeutic target for ...TWIK-related halothane-inhibited potassium channel (THIK1) maintains the resting membrane potential and regulates potassium efflux in microglia. It is a potential therapeutic target for neurodegenerative disorders, neuropathic pain and inflammation. However, the mechanism underlying its function remains unclear. Here we used cryo-electron microscopy to solve the structures of full-length human THIK1, revealing two inner gates and a C-type selectivity filter gate, distinct from other two-pore-domain potassium channels. One inner gate, formed by a short helix in the distal C terminus, introduces a unique gating mechanism involving the distal cytoplasmic domain. The other, beneath the selectivity filter, is constricted by Y273 in the M4 helix, dividing the cavity. In addition, the selectivity filter gate is modulated by polyunsaturated fatty acids. These structural insights into THIK1 gating, through the distal C-terminal helices, hydrophilic residues and selectivity filter, advance our understanding of THIK1's role in microglial homeostasis and neuropathologies. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61467.map.gz emd_61467.map.gz | 27.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61467-v30.xml emd-61467-v30.xml emd-61467.xml emd-61467.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61467.png emd_61467.png | 24 KB | ||
| Filedesc metadata |  emd-61467.cif.gz emd-61467.cif.gz | 6.3 KB | ||
| Others |  emd_61467_half_map_1.map.gz emd_61467_half_map_1.map.gz emd_61467_half_map_2.map.gz emd_61467_half_map_2.map.gz | 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61467 http://ftp.pdbj.org/pub/emdb/structures/EMD-61467 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61467 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61467 | HTTPS FTP |
-Related structure data
| Related structure data |  9jgzMC  9jh1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61467.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61467.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61467_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61467_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Wild type K2P ion channel
| Entire | Name: Wild type K2P ion channel |
|---|---|
| Components |
|
-Supramolecule #1: Wild type K2P ion channel
| Supramolecule | Name: Wild type K2P ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium channel subfamily K member 13
| Macromolecule | Name: Potassium channel subfamily K member 13 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.027766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGHLNEDNA RFLLLAALIV LYLLGGAAVF SALELAHERQ AKQRWEERLA NFSRGHNLSR DELRGFLRHY EEATRAGIRV DNVRPRWDF TGAFYFVGTV VSTIGFGMTT PATVGGKIFL IFYGLVGCSS TILFFNLFLE RLITIIAYIM KSCHQRQLRR R GALPQESL ...String: GPGHLNEDNA RFLLLAALIV LYLLGGAAVF SALELAHERQ AKQRWEERLA NFSRGHNLSR DELRGFLRHY EEATRAGIRV DNVRPRWDF TGAFYFVGTV VSTIGFGMTT PATVGGKIFL IFYGLVGCSS TILFFNLFLE RLITIIAYIM KSCHQRQLRR R GALPQESL KDAGQCEVDS LAGWKPSVYY VMLILCTASI LISCCASAMY TPIEGWSYFD SLYFCFVAFS TIGFGDLVSS QN AHYESQG LYRFANFVFI LMGVCCIYSL FNVISILIKQ SLNWILRKMD SGCCPQCQRG LLRSRRNVVM PGSVRNRCNI SIE TDGVAE SDTDGRRLSG EMISMKDLLA ANKASLAILQ KQLSEMANGC PHQTSTLARD NEFSGGVGAF AIMNNRLAE UniProtKB: Potassium channel subfamily K member 13 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 12 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: LINOLEIC ACID
| Macromolecule | Name: LINOLEIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: EIC |
|---|---|
| Molecular weight | Theoretical: 280.445 Da |
| Chemical component information |  ChemComp-EIC: |
-Macromolecule #4: ARACHIDONIC ACID
| Macromolecule | Name: ARACHIDONIC ACID / type: ligand / ID: 4 / Number of copies: 2 / Formula: ACD |
|---|---|
| Molecular weight | Theoretical: 304.467 Da |
| Chemical component information | 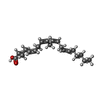 ChemComp-ACD: |
-Macromolecule #5: (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec...
| Macromolecule | Name: (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate type: ligand / ID: 5 / Number of copies: 2 / Formula: D21 |
|---|---|
| Molecular weight | Theoretical: 674.929 Da |
| Chemical component information |  ChemComp-D21: |
-Macromolecule #6: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9jgz: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































