[English] 日本語
 Yorodumi
Yorodumi- EMDB-61433: Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Uroten... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Urotensin receptor (UTS2R)-Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / GPCR / Urotensin receptor / UTS2R / Gq / agonist / [Pen5]-urotensin (4-11) / complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationurotensin II receptor activity / blood circulation / neuropeptide signaling pathway / Peptide ligand-binding receptors / blood vessel diameter maintenance / G protein-coupled receptor activity / regulation of blood pressure / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta ...urotensin II receptor activity / blood circulation / neuropeptide signaling pathway / Peptide ligand-binding receptors / blood vessel diameter maintenance / G protein-coupled receptor activity / regulation of blood pressure / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Xu HE / You C / Gao T / Duan J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2025 Journal: J Biol Chem / Year: 2025Title: Structural insights into hormone recognition and G-protein coupling of the urotensin-II receptor. Authors: Tianyu Gao / Chongzhao You / Yinglong Cao / Xiaofang Xu / Qingning Yuan / Shiyi Shen / H Eric Xu / Jia Duan /  Abstract: Urotensin-II (U-II) is a potent vasoconstrictor peptide that interacts with the human urotensin-II receptor (UTR), a class A G protein-coupled receptor (GPCR) that primarily couples with G proteins. ...Urotensin-II (U-II) is a potent vasoconstrictor peptide that interacts with the human urotensin-II receptor (UTR), a class A G protein-coupled receptor (GPCR) that primarily couples with G proteins. In this study, we present the cryo-electron microscopy structure of the miniG-coupled UTR bound to the potent UTR agonist P5U, providing insights into unique ligand recognition and activation mechanisms. Unlike typical linear peptides, the cyclic structure of P5U engages the receptor's transmembrane domains through key side chain interactions involving residues F6, W7, K8, and Y9, which are crucial for receptor activation. Comparative analysis with somatostatin receptors (SSTRs) reveals distinct ligand specificity, driven by variations in side chain composition. Notably, we identify F274 as the toggle switch residue in UTR, in contrast to the classical W seen in other GPCRs. Our findings elucidate the structural basis for UTR's G coupling specificity, highlighting unique Gα interactions. This study advances the understanding of U-II signaling and offers a foundation for developing selective UTR modulators, with potential therapeutic implications for cardiovascular diseases linked to dysregulated U-II activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61433.map.gz emd_61433.map.gz | 32.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61433-v30.xml emd-61433-v30.xml emd-61433.xml emd-61433.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61433.png emd_61433.png | 22.9 KB | ||
| Filedesc metadata |  emd-61433.cif.gz emd-61433.cif.gz | 6.5 KB | ||
| Others |  emd_61433_half_map_1.map.gz emd_61433_half_map_1.map.gz emd_61433_half_map_2.map.gz emd_61433_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61433 http://ftp.pdbj.org/pub/emdb/structures/EMD-61433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61433 | HTTPS FTP |
-Related structure data
| Related structure data |  9jfkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61433.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61433.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_61433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Uroten...
| Entire | Name: Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Urotensin receptor (UTS2R)-Gq complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Uroten...
| Supramolecule | Name: Cryo-EM structure of [Pen5]-urotensin (4-11)-bounded human Urotensin receptor (UTS2R)-Gq complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #2: G subunit q (gi2-mini-gq chimeric)
| Macromolecule | Name: G subunit q (gi2-mini-gq chimeric) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.084832 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA ...String: MGSTVSAEDK AAAERSKMID KNLREDGEKA RRTLRLLLLG ADNSGKSTIV KQMRILHGGS GGSGGTSGIF ETKFQVDKVN FHMFDVGGQ RDERRKWIQC FNDVTAIIFV VDSSDYNRLQ EALNDFKSIW NNRWLRTISV ILFLNKQDLL AEKVLAGKSK I EDYFPEFA RYTTPEDATP EPGEDPRVTR AKYFIRKEFV DISTASGDGR HICYPHFTCA VDTENARRIF NDCKDIILQM NL REYNLV |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nano body 35
| Macromolecule | Name: Nano body 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Macromolecule #5: Urotensin-2 receptor
| Macromolecule | Name: Urotensin-2 receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.857465 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MALTPESPSS FPGLAATGSS VPEPPGGPNA TLNSSWASPT EPSSLEDLVA TGTIGTLLSA MGVVGVVGNA YTLVVTCRSL RAVASMYVY VVNLALADLL YLLSIPFIVA TYVTKEWHFG DVGCRVLFGL DFLTMHASIF TLTVMSSERY AAVLRPLDTV Q RPKGYRKL ...String: MALTPESPSS FPGLAATGSS VPEPPGGPNA TLNSSWASPT EPSSLEDLVA TGTIGTLLSA MGVVGVVGNA YTLVVTCRSL RAVASMYVY VVNLALADLL YLLSIPFIVA TYVTKEWHFG DVGCRVLFGL DFLTMHASIF TLTVMSSERY AAVLRPLDTV Q RPKGYRKL LALGTWLLAL LLTLPVMLAM RLVRRGPKSL CLPAWGPRAH RAYLTLLFAT SIAGPGLLIG LLYARLARAY RR SQRASFK RARRPGARAL RLVLGIVLLF WACFLPFWLW QLLAQYHQAP LAPRTARIVN YLTTCLTYGN SCANPFLYTL LTR NYRDHL RGRVRGPGSG GGRGPVPSLQ P UniProtKB: Urotensin-2 receptor |
-Macromolecule #6: ASP-LE1-PHE-TRP-LYS-TYR-CYS-VAL
| Macromolecule | Name: ASP-LE1-PHE-TRP-LYS-TYR-CYS-VAL / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.09231 KDa |
| Sequence | String: D(LE1)FWKYCV |
-Macromolecule #7: GLYCOCHENODEOXYCHOLIC ACID
| Macromolecule | Name: GLYCOCHENODEOXYCHOLIC ACID / type: ligand / ID: 7 / Number of copies: 2 / Formula: CHO |
|---|---|
| Molecular weight | Theoretical: 449.623 Da |
| Chemical component information | 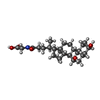 ChemComp-CHO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 30.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































