+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TauT | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein structure / STRUCTUAL PROTEIN / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtaurine transmembrane transporter activity / alanine transmembrane transporter activity / gamma-aminobutyric acid transmembrane transporter activity / taurine:sodium symporter activity / import across plasma membrane / gamma-aminobutyric acid import / gamma-aminobutyric acid:sodium:chloride symporter activity / alanine transport / amino acid:sodium symporter activity / taurine transmembrane transport ...taurine transmembrane transporter activity / alanine transmembrane transporter activity / gamma-aminobutyric acid transmembrane transporter activity / taurine:sodium symporter activity / import across plasma membrane / gamma-aminobutyric acid import / gamma-aminobutyric acid:sodium:chloride symporter activity / alanine transport / amino acid:sodium symporter activity / taurine transmembrane transport / amino acid import across plasma membrane / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / SLC-mediated transport of neurotransmitters / neurotransmitter transport / microvillus membrane / amino acid transport / transport across blood-brain barrier / cell projection / sodium ion transmembrane transport / positive regulation of cell differentiation / modulation of chemical synaptic transmission / GABA-ergic synapse / basolateral plasma membrane / postsynaptic membrane / apical plasma membrane / neuronal cell body / dendrite / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Zhao Y / Xu H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2025 Journal: Cell Discov / Year: 2025Title: Structural characterization reveals substrate recognition by the taurine transporter TauT. Authors: Hao Xu / Qinru Bai / Han Wang / Jun Zhao / Aiping Guo / Renjie Li / Qihao Chen / Yiqing Wei / Na Li / Zhuo Huang / Yan Zhao /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61208.map.gz emd_61208.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61208-v30.xml emd-61208-v30.xml emd-61208.xml emd-61208.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61208.png emd_61208.png | 40.6 KB | ||
| Filedesc metadata |  emd-61208.cif.gz emd-61208.cif.gz | 6.3 KB | ||
| Others |  emd_61208_half_map_1.map.gz emd_61208_half_map_1.map.gz emd_61208_half_map_2.map.gz emd_61208_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61208 http://ftp.pdbj.org/pub/emdb/structures/EMD-61208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61208 | HTTPS FTP |
-Related structure data
| Related structure data |  9j7nMC  9j7mC  9j7oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61208.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61208.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_61208_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61208_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : structure of TauT
| Entire | Name: structure of TauT |
|---|---|
| Components |
|
-Supramolecule #1: structure of TauT
| Supramolecule | Name: structure of TauT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium- and chloride-dependent taurine transporter
| Macromolecule | Name: Sodium- and chloride-dependent taurine transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.881633 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATKEKLQCL KDFHKDILKP SPGKSPGTRP EDEAEGKPPQ REKWSSKIDF VLSVAGGFVG LGNVWRFPYL CYKNGGGAFL IPYFIFLFG SGLPVFFLEI IIGQYTSEGG ITCWEKICPL FSGIGYASVV IVSLLNVYYI VILAWATYYL FQSFQKELPW A HCNHSWNT ...String: MATKEKLQCL KDFHKDILKP SPGKSPGTRP EDEAEGKPPQ REKWSSKIDF VLSVAGGFVG LGNVWRFPYL CYKNGGGAFL IPYFIFLFG SGLPVFFLEI IIGQYTSEGG ITCWEKICPL FSGIGYASVV IVSLLNVYYI VILAWATYYL FQSFQKELPW A HCNHSWNT PHCMEDTMRK NKSVWITISS TNFTSPVIEF WERNVLSLSP GIDHPGSLKW DLALCLLLVW LVCFFCIWKG VR STGKVVY FTATFPFAML LVLLVRGLTL PGAGAGIKFY LYPDITRLED PQVWIDAGTQ IFFSYAICLG AMTSLGSYNK YKY NSYRDC MLLGCLNSGT SFVSGFAIFS ILGFMAQEQG VDIADVAESG PGLAFIAYPK AVTMMPLPTF WSILFFIMLL LLGL DSQFV EVEGQITSLV DLYPSFLRKG YRREIFIAFV CSISYLLGLT MVTEGGMYVF QLFDYYAASG VCLLWVAFFE CFVIA WIYG GDNLYDGIED MIGYRPGPWM KYSWAVITPV LCVGCFIFSL VKYVPLTYNK TYVYPNWAIG LGWSLALSSM LCVPLV IVI RLCQTEGPFL VRVKYLLTPR EPNRWAVERE GATPYNSRTV MNGALVKPTH IIVETMM UniProtKB: Sodium- and chloride-dependent taurine transporter |
-Macromolecule #2: BETA-ALANINE
| Macromolecule | Name: BETA-ALANINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: BAL |
|---|---|
| Molecular weight | Theoretical: 89.093 Da |
| Chemical component information | 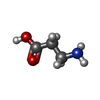 ChemComp-BAL: |
-Macromolecule #3: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 3 / Number of copies: 8 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































