+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the Planar Cell Polarity Core Protein Vangl1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NTD / PCP / Vangl / Prickle / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmucociliary clearance / RHOD GTPase cycle / RHOF GTPase cycle / RND1 GTPase cycle / Wnt signaling pathway, planar cell polarity pathway / RND2 GTPase cycle / RND3 GTPase cycle / RHOB GTPase cycle / RHOV GTPase cycle / pigmentation ...mucociliary clearance / RHOD GTPase cycle / RHOF GTPase cycle / RND1 GTPase cycle / Wnt signaling pathway, planar cell polarity pathway / RND2 GTPase cycle / RND3 GTPase cycle / RHOB GTPase cycle / RHOV GTPase cycle / pigmentation / RHOC GTPase cycle / RHOJ GTPase cycle / RHOQ GTPase cycle / RHOU GTPase cycle / CDC42 GTPase cycle / RHOH GTPase cycle / lateral plasma membrane / RHOG GTPase cycle / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / RAC1 GTPase cycle / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Zhang F / Chen S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Cryo-EM structure and oligomerization of the human planar cell polarity core protein Vangl1. Authors: Fan Zhang / Shaobai Li / Hao Wu / Shanshuang Chen /  Abstract: Vangl is a planar cell polarity (PCP) core protein essential for aligned cell orientation along the epithelial plane perpendicular to the apical-basal direction, which is important for tissue ...Vangl is a planar cell polarity (PCP) core protein essential for aligned cell orientation along the epithelial plane perpendicular to the apical-basal direction, which is important for tissue morphogenesis, development and collective cell behavior. Mutations in Vangl are associated with developmental defects, including neural tube defects (NTDs), according to human cohort studies of sporadic and familial cases. The complex mechanisms underlying Vangl-mediated PCP signaling or Vangl-associated human congenital diseases have been hampered by the lack of molecular characterizations of Vangl. Here, we show biochemical and structural evidence that human Vangl1 oligomerizes as dimers of trimers, and that the dimerization of trimers promotes binding to the PCP effector Prickle1 (Pk1) in vitro. Mapping of human disease-associated point mutations suggests potential pathological mechanisms and paves the way for future studies on the importance of lipid binding, central vestibule and oligomerization of Vangl, thereby providing insights into the molecular mechanisms of the PCP signaling pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60540.map.gz emd_60540.map.gz | 64.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60540-v30.xml emd-60540-v30.xml emd-60540.xml emd-60540.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60540.png emd_60540.png | 43.9 KB | ||
| Filedesc metadata |  emd-60540.cif.gz emd-60540.cif.gz | 5.9 KB | ||
| Others |  emd_60540_half_map_1.map.gz emd_60540_half_map_1.map.gz emd_60540_half_map_2.map.gz emd_60540_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60540 http://ftp.pdbj.org/pub/emdb/structures/EMD-60540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60540 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60540 | HTTPS FTP |
-Validation report
| Summary document |  emd_60540_validation.pdf.gz emd_60540_validation.pdf.gz | 753 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60540_full_validation.pdf.gz emd_60540_full_validation.pdf.gz | 752.5 KB | Display | |
| Data in XML |  emd_60540_validation.xml.gz emd_60540_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_60540_validation.cif.gz emd_60540_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60540 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60540 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60540 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60540 | HTTPS FTP |
-Related structure data
| Related structure data |  8zxdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60540.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60540.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60540_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_60540_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vang-like protein 1
| Entire | Name: Vang-like protein 1 |
|---|---|
| Components |
|
-Supramolecule #1: Vang-like protein 1
| Supramolecule | Name: Vang-like protein 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Vang-like protein 1
| Macromolecule | Name: Vang-like protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.053887 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDTESTYSGY SYYSSHSKKS HRQGERTRER HKSPRNKDGR GSEKSVTIQP PTGEPLLGND STRTEEVQDD NWGETTTAIT GTSEHSISQ EDIARISKDM EDSVGLDCKR YLGLTVASFL GLLVFLTPIA FILLPPILWR DELEPCGTIC EGLFISMAFK L LILLIGTW ...String: MDTESTYSGY SYYSSHSKKS HRQGERTRER HKSPRNKDGR GSEKSVTIQP PTGEPLLGND STRTEEVQDD NWGETTTAIT GTSEHSISQ EDIARISKDM EDSVGLDCKR YLGLTVASFL GLLVFLTPIA FILLPPILWR DELEPCGTIC EGLFISMAFK L LILLIGTW ALFFRKRRAD MPRVFVFRAL LLVLIFLFVV SYWLFYGVRI LDSRDRNYQG IVQYAVSLVD ALLFIHYLAI VL LELRQLQ PMFTLQVVRS TDGESRFYSL GHLSIQRAAL VVLENYYKDF TIYNPNLLTA SKFRAAKHMA GLKVYNVDGP SNN ATGQSR AMIAAAARRR DSSHNELYYE EAEHERRVKK RKARLVVAVE EAFIHIQRLQ AEEQQKAPGE VMDPREAAQA IFPS MARAL QKYLRITRQQ NYHSMESILQ HLAFCITNGM TPKAFLERYL SAGPTLQYDK DRWLSTQWRL VSDEAVTNGL RDGIV FVLK CLDFSLVVNV KKIPFIILSE EFIDPKSHKF VLRLQSETSV UniProtKB: Vang-like protein 1 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 6 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information | 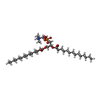 ChemComp-MC3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































