+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the apo hTAAR1-Gs complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / TAAR1 / Cryo-EM / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationAmine ligand-binding receptors / trace-amine receptor activity / endomembrane system / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta ...Amine ligand-binding receptors / trace-amine receptor activity / endomembrane system / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / endoplasmic reticulum membrane / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Jiang KX / Zheng Y / Xu F | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules. Authors: Kexin Jiang / You Zheng / Liting Zeng / Ling Wang / Fei Li / Jun Pu / Yingli Lu / Suwen Zhao / Fei Xu /  Abstract: Increasing global concerns about psychoactive substance addiction and psychotic disorders highlight the need for comprehensive research into the structure-function relationship governing ligand ...Increasing global concerns about psychoactive substance addiction and psychotic disorders highlight the need for comprehensive research into the structure-function relationship governing ligand recognition between these substances and their receptors in the brain. Recent studies indicate the significant involvement of trace amine-associated receptor 1 (TAAR1) in the signaling regulation of the hallucinogen lysergic acid diethylamide (LSD) and other antipsychotic drugs. This study presents structures of the TAAR1-Gs protein complex recognizing LSD, which exhibits a polypharmacological profile, and the partial agonist RO5263397, which is a drug candidate for schizophrenia and addiction. Moreover, we elucidate the cross-species recognition and partial activation mechanism for TAAR1, which holds promising implications from a drug discovery perspective. Through mutagenesis, functional studies, and molecular dynamics (MD) simulations, we provide a comprehensive understanding of a versatile TAAR1 pocket in recognizing various ligands as well as in the ligand-free state, underpinning the structural basis of its high adaptability. These findings offer valuable insights for the design of antipsychotic drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60417.map.gz emd_60417.map.gz | 53.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60417-v30.xml emd-60417-v30.xml emd-60417.xml emd-60417.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60417.png emd_60417.png | 73.4 KB | ||
| Filedesc metadata |  emd-60417.cif.gz emd-60417.cif.gz | 6.9 KB | ||
| Others |  emd_60417_half_map_1.map.gz emd_60417_half_map_1.map.gz emd_60417_half_map_2.map.gz emd_60417_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60417 http://ftp.pdbj.org/pub/emdb/structures/EMD-60417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60417 | HTTPS FTP |
-Validation report
| Summary document |  emd_60417_validation.pdf.gz emd_60417_validation.pdf.gz | 905.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60417_full_validation.pdf.gz emd_60417_full_validation.pdf.gz | 905.3 KB | Display | |
| Data in XML |  emd_60417_validation.xml.gz emd_60417_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_60417_validation.cif.gz emd_60417_validation.cif.gz | 15.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60417 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60417 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60417 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60417 | HTTPS FTP |
-Related structure data
| Related structure data |  8zsjMC  8zspC  8zssC  8zsvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60417.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60417.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
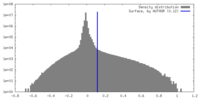
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60417_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
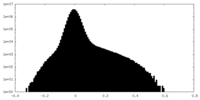
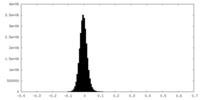
| Density Histograms |
-Half map: #2
| File | emd_60417_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
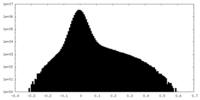
| Density Histograms |
- Sample components
Sample components
-Entire : The apo hTAAR1-Gs complex
| Entire | Name: The apo hTAAR1-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: The apo hTAAR1-Gs complex
| Supramolecule | Name: The apo hTAAR1-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.359992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDGEKA AAATHRLLLL GADNSGKSTI VKQMRIYHGG SGGSGVNSGI FETKFQVDKV NFHMFDVGA QRDERRKWIQ CFNDVTAIIF VVDSSDYNRL QEALNDFKSI WNNRWLRTIS VILFLNKQDL LAEKVLAGKS K IEDYFPEF ...String: MGSTVSAEDK AAAERSKMID KNLREDGEKA AAATHRLLLL GADNSGKSTI VKQMRIYHGG SGGSGVNSGI FETKFQVDKV NFHMFDVGA QRDERRKWIQ CFNDVTAIIF VVDSSDYNRL QEALNDFKSI WNNRWLRTIS VILFLNKQDL LAEKVLAGKS K IEDYFPEF ARYTTPEDAT PEPGEDPRVT RAKYFIRDEF LRISTASGDG RHYCYPHFTC SVDTENARRI FNDCRDIIQR MH LRQYELL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.271938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQVQLQESGG GLVQPGGSLR LSCAASGFTF SNYKMNWVRQ APGKGLEWVS DISQSGASIS YTGSVKGRFT ISRDNAKNTL YLQMNSLKP EDTAVYYCAR CPAPFTRDCF DVTSTTYAYR GQGTQVTVSS HHHHHHEPEA |
-Macromolecule #5: Soluble cytochrome b562,Trace amine-associated receptor 1
| Macromolecule | Name: Soluble cytochrome b562,Trace amine-associated receptor 1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.645188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLM GQPGNGSAMP F CHNIINIS ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLM GQPGNGSAMP F CHNIINIS CVKNNWSNDV RASLYSLMVL IILTTLVGNL IVIVSISHFK QLHTPTNWLI HSMATVDFLL GCLVMPYSMV RS AEHCWYF GEVFCKIHTS TDIMLSSASI FHLSFISIDR YYAVCDPLRY KAKMNILVIC VMIFISWSVP AVFAFGMIFL ELN FKGAEE IYYKHVHCRG GCSVFFSKIS GVLTFMTSFY IPGSIMLCVY YRIYLIAKEQ ARLISDANQK LQIGLEMKNG ISQS KERKA VKTLGIVMGV FLICWCPFFI CTVMDPFLHY IIPPTLNDVL IWFGYLNSTF NPMVYAFFYP WFRKALKMML FGKIF QKDS SRCKLFLELS S UniProtKB: Soluble cytochrome b562, Trace amine-associated receptor 1 |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)