[English] 日本語
 Yorodumi
Yorodumi- EMDB-60404: Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-DNGI complex | ||||||||||||
 Map data Map data | full map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ENDOTHELIN / RECEPTOR / Gi / COMPLEX / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationenteric smooth muscle cell differentiation / response to endothelin / : / negative regulation of neuron maturation / endothelin A receptor binding / chordate pharynx development / rhythmic excitation / neuroblast migration / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity ...enteric smooth muscle cell differentiation / response to endothelin / : / negative regulation of neuron maturation / endothelin A receptor binding / chordate pharynx development / rhythmic excitation / neuroblast migration / negative regulation of phospholipase C/protein kinase C signal transduction / endothelin receptor activity / peptide hormone secretion / endothelin B receptor binding / aldosterone metabolic process / cellular response to human chorionic gonadotropin stimulus / meiotic cell cycle process involved in oocyte maturation / semaphorin-plexin signaling pathway involved in axon guidance / positive regulation of artery morphogenesis / histamine secretion / neural crest cell fate commitment / regulation of fever generation / vein smooth muscle contraction / glomerular endothelium development / response to prostaglandin F / sympathetic neuron axon guidance / positive regulation of penile erection / positive regulation of sarcomere organization / noradrenergic neuron differentiation / positive regulation of chemokine-mediated signaling pathway / leukocyte activation / phospholipase D-activating G protein-coupled receptor signaling pathway / maternal process involved in parturition / rough endoplasmic reticulum lumen / posterior midgut development / body fluid secretion / pharyngeal arch artery morphogenesis / regulation of D-glucose transmembrane transport / endothelin receptor signaling pathway involved in heart process / positive regulation of odontogenesis / epithelial fluid transport / cardiac neural crest cell migration involved in outflow tract morphogenesis / heparin proteoglycan metabolic process / negative regulation of hormone secretion / response to ozone / Weibel-Palade body / podocyte differentiation / endothelin receptor signaling pathway / positive regulation of cation channel activity / developmental pigmentation / positive regulation of cell growth involved in cardiac muscle cell development / renal sodium excretion / response to sodium phosphate / response to leptin / axonogenesis involved in innervation / glomerular filtration / enteric nervous system development / renin secretion into blood stream / positive regulation of smooth muscle contraction / renal sodium ion absorption / renal albumin absorption / cellular response to follicle-stimulating hormone stimulus / artery smooth muscle contraction / positive regulation of prostaglandin secretion / protein transmembrane transport / cellular response to luteinizing hormone stimulus / regulation of pH / cellular response to mineralocorticoid stimulus / respiratory gaseous exchange by respiratory system / melanocyte differentiation / vasoconstriction / positive regulation of renal sodium excretion / basal part of cell / peripheral nervous system development / type 1 angiotensin receptor binding / response to salt / negative regulation of adenylate cyclase activity / positive regulation of hormone secretion / regulation of systemic arterial blood pressure by endothelin / regulation of epithelial cell proliferation / dorsal/ventral pattern formation / cellular response to toxic substance / embryonic heart tube development / axon extension / cellular response to fatty acid / cartilage development / establishment of endothelial barrier / prostaglandin biosynthetic process / positive regulation of neutrophil chemotaxis / positive regulation of urine volume / signal transduction involved in regulation of gene expression / superoxide anion generation / negative regulation of protein metabolic process / neural crest cell migration / : / cellular response to glucocorticoid stimulus / nitric oxide transport / response to pain / middle ear morphogenesis / branching involved in blood vessel morphogenesis / response to dexamethasone / positive regulation of cardiac muscle hypertrophy Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
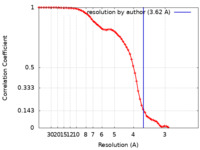
| Method | single particle reconstruction / cryo EM / Resolution: 3.62 Å | ||||||||||||
 Authors Authors | Tani K / Maki-Yonekura S / Kanno R / Negami T / Hamaguchi T / Hall M / Mizoguchi A / Humbel BM / Terada T / Yonekura K / Doi T | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Structure of endothelin ET receptor-G complex in a conformation stabilized by unique NPxxL motif. Authors: Kazutoshi Tani / Saori Maki-Yonekura / Ryo Kanno / Tatsuki Negami / Tasuku Hamaguchi / Malgorzata Hall / Akira Mizoguchi / Bruno M Humbel / Tohru Terada / Koji Yonekura / Tomoko Doi /  Abstract: Endothelin type B receptor (ETR) plays a crucial role in regulating blood pressure and humoral homeostasis, making it an important therapeutic target for related diseases. ETR activation by the ...Endothelin type B receptor (ETR) plays a crucial role in regulating blood pressure and humoral homeostasis, making it an important therapeutic target for related diseases. ETR activation by the endogenous peptide hormones endothelin (ET)-1-3 stimulates several signaling pathways, including G, G, G, G, and β-arrestin. Although the conserved NPxxY motif in transmembrane helix 7 (TM7) is important during GPCR activation, ETR possesses the lesser known NPxxL motif. In this study, we present the cryo-EM structure of the ETR-G complex, complemented by MD simulations and functional studies. These investigations reveal an unusual movement of TM7 to the intracellular side during ETR activation and the essential roles of the diverse NPxxL motif in stabilizing the active conformation of ETR and organizing the assembly of the binding pocket for the α5 helix of G protein. These findings enhance our understanding of the interactions between GPCRs and G proteins, thereby advancing the development of therapeutic strategies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60404.map.gz emd_60404.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60404-v30.xml emd-60404-v30.xml emd-60404.xml emd-60404.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60404_fsc.xml emd_60404_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_60404.png emd_60404.png | 79.3 KB | ||
| Filedesc metadata |  emd-60404.cif.gz emd-60404.cif.gz | 6.1 KB | ||
| Others |  emd_60404_half_map_1.map.gz emd_60404_half_map_1.map.gz emd_60404_half_map_2.map.gz emd_60404_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60404 http://ftp.pdbj.org/pub/emdb/structures/EMD-60404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60404 | HTTPS FTP |
-Related structure data
| Related structure data |  8zrtMC  8xwpC  8xwqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60404.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60404.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.19 Å | ||||||||||||||||||||||||||||||||||||
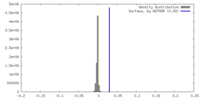
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half-odd map
| File | emd_60404_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-odd map | ||||||||||||
| Projections & Slices |
| ||||||||||||
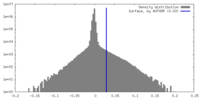
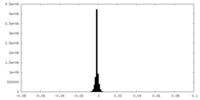
| Density Histograms |
-Half map: half-even map
| File | emd_60404_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-even map | ||||||||||||
| Projections & Slices |
| ||||||||||||
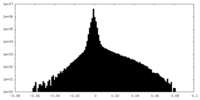
| Density Histograms |
- Sample components
Sample components
-Entire : ET-1 BOUND ETBR COMPLEX
| Entire | Name: ET-1 BOUND ETBR COMPLEX |
|---|---|
| Components |
|
-Supramolecule #1: ET-1 BOUND ETBR COMPLEX
| Supramolecule | Name: ET-1 BOUND ETBR COMPLEX / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endothelin receptor type B
| Macromolecule | Name: Endothelin receptor type B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.808262 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PGGGLAPAEV PKGDRTAGSP PRTISPPPCQ GPIEIKETFK YINTVVSCLV FVLGIIGNST LLYIIYKNKC MRNGPNILIA SLALGDLLH IVIDIPINVY KLLAEDWPFG AEMCKLVPFI QKASVGITVL SLCALSIDRY RAVASWSRIK GIGVPKWTAV E IVLIWVVS ...String: PGGGLAPAEV PKGDRTAGSP PRTISPPPCQ GPIEIKETFK YINTVVSCLV FVLGIIGNST LLYIIYKNKC MRNGPNILIA SLALGDLLH IVIDIPINVY KLLAEDWPFG AEMCKLVPFI QKASVGITVL SLCALSIDRY RAVASWSRIK GIGVPKWTAV E IVLIWVVS VVLAVPEAIG FDIITMDYKG SYLRICLLHP VQKTAFMQFY KTAKDWWLFS FYFCLPLAIT AFFYTLMTCE ML RKKSGMQ IALNDHLKQR REVAKTVFCL VLVFALCWLP LHLSRILKLT LYNQNDPNRC ELLSFLLVLD YIGINMASLN SCI NPIALY LVSKRFKNAF KSALCCWAQS UniProtKB: Endothelin receptor type B |
-Macromolecule #2: Endothelin-1
| Macromolecule | Name: Endothelin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.497951 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSCSSLMDKE CVYFCHLDII W UniProtKB: Endothelin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 3.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8zrt: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)