+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Anaerobically isolated active [FeFe]-hydrogenase CbA5H | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | [FeFe]-hydrogenases / homodimer / Zn-binding / OXIDOREDUCTASE | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationferredoxin hydrogenase activity / 4 iron, 4 sulfur cluster binding / iron ion binding Similarity search - Function | |||||||||||||||||||||
| Biological species |  Clostridium beijerinckii (bacteria) Clostridium beijerinckii (bacteria) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.21 Å | |||||||||||||||||||||
 Authors Authors | Kawamoto A / Kurisu G | |||||||||||||||||||||
| Funding support |  Germany, Germany,  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural determinants of oxygen resistance and Zn-mediated stability of the [FeFe]-hydrogenase from . Authors: Jifu Duan / Andreas Rutz / Akihiro Kawamoto / Shuvankar Naskar / Kristina Edenharter / Silke Leimkühler / Eckhard Hofmann / Thomas Happe / Genji Kurisu /   Abstract: [FeFe]-hydrogenases catalyze the reversible two-electron reduction of two protons to molecular hydrogen. Although these enzymes are among the most efficient H-converting biocatalysts in nature, their ...[FeFe]-hydrogenases catalyze the reversible two-electron reduction of two protons to molecular hydrogen. Although these enzymes are among the most efficient H-converting biocatalysts in nature, their catalytic cofactor (termed H-cluster) is irreversibly destroyed upon contact with dioxygen. The [FeFe]-hydrogenase CbA5H from has a unique mechanism to protect the H-cluster from oxygen-induced degradation. The protective strategy of CbA5H was proposed based on a partial protein structure of CbA5H's oxygen-shielded form. Here, we present a cryo-EM structure of 2.2 Å resolution from the entire enzyme in its dimeric and active state and elucidate the structural parameters of the reversible cofactor protection mechanism. We found that both subunits of the homodimeric structure of CbA5H have a Zn-binding four-helix domain, which does not play a role in electron transport as described for other complex protein structures. Biochemical data instead confirm that two [4Fe-4S] clusters are responsible for electron transfer in CbA5H, while the identified zinc atom is critical for oligomerization and protein stability. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60383.map.gz emd_60383.map.gz | 10.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60383-v30.xml emd-60383-v30.xml emd-60383.xml emd-60383.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60383_fsc.xml emd_60383_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_60383.png emd_60383.png | 82.8 KB | ||
| Masks |  emd_60383_msk_1.map emd_60383_msk_1.map | 107.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60383.cif.gz emd-60383.cif.gz | 7.3 KB | ||
| Others |  emd_60383_additional_1.map.gz emd_60383_additional_1.map.gz emd_60383_half_map_1.map.gz emd_60383_half_map_1.map.gz emd_60383_half_map_2.map.gz emd_60383_half_map_2.map.gz | 82.6 MB 82.8 MB 82.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60383 http://ftp.pdbj.org/pub/emdb/structures/EMD-60383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60383 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60383 | HTTPS FTP |
-Related structure data
| Related structure data |  8zqdMC  9f46C  9f47C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60383.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60383.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60383_msk_1.map emd_60383_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_60383_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_60383_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60383_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homodimeric structure of the active [FeFe]-hydrogenase CbA5H
| Entire | Name: homodimeric structure of the active [FeFe]-hydrogenase CbA5H |
|---|---|
| Components |
|
-Supramolecule #1: homodimeric structure of the active [FeFe]-hydrogenase CbA5H
| Supramolecule | Name: homodimeric structure of the active [FeFe]-hydrogenase CbA5H type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Clostridium beijerinckii (bacteria) Clostridium beijerinckii (bacteria) |
-Macromolecule #1: [FeFe]-hydrogenase
| Macromolecule | Name: [FeFe]-hydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium beijerinckii (bacteria) Clostridium beijerinckii (bacteria) |
| Molecular weight | Theoretical: 75.001938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDNKKSFIQ SALGSVFSVF SEEELKELSN GRKIAICGKV NNPGIIEVPE GATLNEIIQL CGGLINKSNF KAAQIGLPFG GFLTEDSLD KEFDFGIFYE NIARTIIVLS QEDCIIQFEK FYIEYLLAKI KDGSYKNYEV VKEDITEMFN ILNRISKGVS N MREIYLLR ...String: MGDNKKSFIQ SALGSVFSVF SEEELKELSN GRKIAICGKV NNPGIIEVPE GATLNEIIQL CGGLINKSNF KAAQIGLPFG GFLTEDSLD KEFDFGIFYE NIARTIIVLS QEDCIIQFEK FYIEYLLAKI KDGSYKNYEV VKEDITEMFN ILNRISKGVS N MREIYLLR NLAVTVKSKM NQKHNIMEEI IDKFYEEIEE HIEEKKCYTS QCNHLVKLTI TKKCIGCGAC KRACPVDCIN GE LKKKHEI DYNRCTHCGA CVSACPVDAI SAGDNTMLFL RDLATPNKVV ITQMAPAVRV AIGEAFGFEP GENVEKKIAA GLR KLGVDY VFDTSWGADL TIMEEAAELQ ERLERHLAGD ESVKLPILTS CCPSWIKFIE QNYGDMLDVP SSAKSPMEMF AIVA KEIWA KEKGLSRDEV TSVAIMPCIA KKYEASRAEF SVDMNYDVDY VITTRELIKI FENSGINLKE IEDEEIDTVM GEYTG AGII FGRTGGVIEA ATRTALEKMT GERFDNIEFE GLRGWDGFRV CELEAGDIKL RIGVAHGLRE AAKMLDKIRS GEEFFH AIE IMACVGGCIG GGGQPKTKGN KQAALQKRAE GLNNIDRSKT LRRSNENPEV LAIYEKYLDH PLSNKAHELL HTVYFPR VK KDDIWSVGVK LFGGGSGGGS GGGSWSHPQF EK UniProtKB: [FeFe]-hydrogenase |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #4: dicarbonyl[bis(cyanide-kappaC)]-mu-(iminodimethanethiolatato-1kap...
| Macromolecule | Name: dicarbonyl[bis(cyanide-kappaC)]-mu-(iminodimethanethiolatato-1kappaS:2kappaS)-mu-(oxomethylidene)diiron(2+) type: ligand / ID: 4 / Number of copies: 2 / Formula: 402 |
|---|---|
| Molecular weight | Theoretical: 354.953 Da |
| Chemical component information | 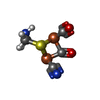 ChemComp-402: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 100 mM Tris-HCl, 2mM NaDT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 50 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: Vitrification carried out in anaerobic condition. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9994 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















































