[English] 日本語
 Yorodumi
Yorodumi- EMDB-60331: Cryo-EM structure of human norepinephrine transporter NET bound w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human norepinephrine transporter NET bound with norepinephrine in an occluded state at a resolution of 2.6 angstrom | |||||||||
 Map data Map data | CryoEM structure of human norepinephrine transporter NET bound with norepinephrine in an occluded state at a resolution of 2.6 angstrom | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | norepinephrine transport / norepinephrine / TRANSPORT PROTEIN/IMMUNE SYSTEM / TRANSPORT PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationneurotransmitter:sodium symporter activity / Defective SLC6A2 causes orthostatic intolerance (OI) / norepinephrine uptake / norepinephrine:sodium symporter activity / norepinephrine transport / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters ...neurotransmitter:sodium symporter activity / Defective SLC6A2 causes orthostatic intolerance (OI) / norepinephrine uptake / norepinephrine:sodium symporter activity / norepinephrine transport / dopamine:sodium symporter activity / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / monoamine transport / SLC-mediated transport of neurotransmitters / response to pain / neuronal cell body membrane / neurotransmitter transport / dopamine uptake involved in synaptic transmission / amino acid transport / alpha-tubulin binding / beta-tubulin binding / sodium ion transmembrane transport / neuron cellular homeostasis / synaptic vesicle membrane / actin binding / presynaptic membrane / chemical synaptic transmission / response to xenobiotic stimulus / axon / cell surface / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Song AL / Wu XD | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Mechanistic insights of substrate transport and inhibitor binding revealed by high-resolution structures of human norepinephrine transporter. Authors: Ailong Song / Xudong Wu /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60331.map.gz emd_60331.map.gz | 78.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60331-v30.xml emd-60331-v30.xml emd-60331.xml emd-60331.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60331.png emd_60331.png | 105.9 KB | ||
| Filedesc metadata |  emd-60331.cif.gz emd-60331.cif.gz | 6.7 KB | ||
| Others |  emd_60331_additional_1.map.gz emd_60331_additional_1.map.gz emd_60331_half_map_1.map.gz emd_60331_half_map_1.map.gz emd_60331_half_map_2.map.gz emd_60331_half_map_2.map.gz | 41.9 MB 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60331 http://ftp.pdbj.org/pub/emdb/structures/EMD-60331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60331 | HTTPS FTP |
-Validation report
| Summary document |  emd_60331_validation.pdf.gz emd_60331_validation.pdf.gz | 783.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60331_full_validation.pdf.gz emd_60331_full_validation.pdf.gz | 783.2 KB | Display | |
| Data in XML |  emd_60331_validation.xml.gz emd_60331_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_60331_validation.cif.gz emd_60331_validation.cif.gz | 15.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60331 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60331 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60331 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60331 | HTTPS FTP |
-Related structure data
| Related structure data |  8zpbMC  8zoyC  8zp1C  8zp2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60331.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60331.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of human norepinephrine transporter NET bound with norepinephrine in an occluded state at a resolution of 2.6 angstrom | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_60331_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap A
| File | emd_60331_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap B
| File | emd_60331_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human norepinephrine transporter NET in complex with Nb_BB4
| Entire | Name: human norepinephrine transporter NET in complex with Nb_BB4 |
|---|---|
| Components |
|
-Supramolecule #1: human norepinephrine transporter NET in complex with Nb_BB4
| Supramolecule | Name: human norepinephrine transporter NET in complex with Nb_BB4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Sodium-dependent noradrenaline transporter
| Macromolecule | Name: Sodium-dependent noradrenaline transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 64.298641 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APRDGDAQPR ETWGKKIDFL LSVVGFAVDL ANVWRFPYLC YKNGGGAFLI PYTLFLIIAG MPLFYMELAL GQYNREGAAT VWKICPFFK GVGYAVILIA LYVGFYYNVI IAWSLYYLFS SFTLNLPWTD CGHTWNSPNC TDPKLLNGSV LGNHTKYSKY K FTPAAEFY ...String: APRDGDAQPR ETWGKKIDFL LSVVGFAVDL ANVWRFPYLC YKNGGGAFLI PYTLFLIIAG MPLFYMELAL GQYNREGAAT VWKICPFFK GVGYAVILIA LYVGFYYNVI IAWSLYYLFS SFTLNLPWTD CGHTWNSPNC TDPKLLNGSV LGNHTKYSKY K FTPAAEFY ERGVLHLHES SGIHDIGLPQ WQLLLCLMVV VIVLYFSLWK GVKTSGKVVW ITATLPYFVL FVLLVHGVTL PG ASNGINA YLHIDFYRLK EATVWIDAAT QIFFSLGAGF GVLIAFASYN KFDNNCYRDA LLTSSINCIT SFVSGFAIFS ILG YMAHEH KVNIEDVATE GAGLVFILYP EAISTLSGST FWAVVFFVML LALGLDSSMG GMEAVITGLA DDFQVLKRHR KLFT FGVTF STFLLALFCI TKGGIYVLTL LDTFAAGTSI LFAVLMEAIG VSWFYGVDRF SNDIQQMMGF RPGLYWRLCW KFVSP AFLL FVVVVSIINF KPLTYDDYIF PPWANWVGWG IALSSMVLVP IYVIYKFLST QGSLWERLAY GITPENEHHL VAQRDI RQF QLQHWLAI UniProtKB: Sodium-dependent noradrenaline transporter |
-Macromolecule #2: Nb_BB4
| Macromolecule | Name: Nb_BB4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.338711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQAGGSLRL SCAASGFPVY QANMYWYRQA PGKEREWVAA IQSEGRTIYA DSVKGRFTIS RDNSKNTVYL QMNSLKPED TAVYYCNVKD AGWASYQYDY WGQGTQVTVS S |
-Macromolecule #3: L-NOREPINEPHRINE
| Macromolecule | Name: L-NOREPINEPHRINE / type: ligand / ID: 3 / Number of copies: 1 / Formula: LNR |
|---|---|
| Molecular weight | Theoretical: 169.178 Da |
| Chemical component information | 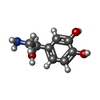 ChemComp-LNR: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 54 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































