[English] 日本語
 Yorodumi
Yorodumi- EMDB-6021: Conformational Spectrum of Multidrug ABC Transporters Revealed by... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6021 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Conformational Spectrum of Multidrug ABC Transporters Revealed by Single Particle Electron Microscopy | |||||||||
 Map data Map data | P-gp in inward-facing conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / P-gp / MsbA | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Moeller A / Chang Lee S / Tao H / Speir S / Chang G / Urbatsch IL / Potter CS / Carragher B / Zhang Q | |||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Distinct conformational spectrum of homologous multidrug ABC transporters. Authors: Arne Moeller / Sung Chang Lee / Houchao Tao / Jeffrey A Speir / Geoffrey Chang / Ina L Urbatsch / Clinton S Potter / Bridget Carragher / Qinghai Zhang /  Abstract: ATP-binding cassette (ABC) exporters are ubiquitously found in all kingdoms of life and their members play significant roles in mediating drug pharmacokinetics and multidrug resistance in the clinic. ...ATP-binding cassette (ABC) exporters are ubiquitously found in all kingdoms of life and their members play significant roles in mediating drug pharmacokinetics and multidrug resistance in the clinic. Significant questions and controversies remain regarding the relevance of their conformations observed in X-ray structures, their structural dynamics, and mechanism of transport. Here, we used single particle electron microscopy (EM) to delineate the entire conformational spectrum of two homologous ABC exporters (bacterial MsbA and mammalian P-glycoprotein) and the influence of nucleotide and substrate binding. Newly developed amphiphiles in complex with lipids that support high protein stability and activity enabled EM visualization of individual complexes in a membrane-mimicking environment. The data provide a comprehensive view of the conformational flexibility of these ABC exporters under various states and demonstrate not only similarities but striking differences between their mechanistic and energetic regulation of conformational changes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6021.map.gz emd_6021.map.gz | 91.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6021-v30.xml emd-6021-v30.xml emd-6021.xml emd-6021.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6021.tif emd_6021.tif | 688.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6021 http://ftp.pdbj.org/pub/emdb/structures/EMD-6021 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6021 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6021 | HTTPS FTP |
-Related structure data
| Related structure data |  6007C  6018C  6019C  6020C  6022C  6023C  6024C  6025C  6026C  6027C  6028C  6029C  6030C  6031C  6032C  6033C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6021.map.gz / Format: CCP4 / Size: 245.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6021.map.gz / Format: CCP4 / Size: 245.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | P-gp in inward-facing conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.455 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
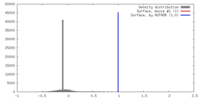
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pgp_m stabilised in lipid/amphiphilic environment - inward-facing...
| Entire | Name: Pgp_m stabilised in lipid/amphiphilic environment - inward-facing 6.0 nm opening |
|---|---|
| Components |
|
-Supramolecule #1000: Pgp_m stabilised in lipid/amphiphilic environment - inward-facing...
| Supramolecule | Name: Pgp_m stabilised in lipid/amphiphilic environment - inward-facing 6.0 nm opening type: sample / ID: 1000 / Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: P-glycoprotein
| Macromolecule | Name: P-glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: P-gp, Pgp, MDR1A / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) / Recombinant strain: Gs115 / Recombinant plasmid: pHIL-mdr3.5-His6 Komagataella pastoris (fungus) / Recombinant strain: Gs115 / Recombinant plasmid: pHIL-mdr3.5-His6 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Staining | Type: NEGATIVE / Details: uranyl formate |
|---|---|
| Grid | Details: thin carbon over holes |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Aug 12, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 456 / Average electron dose: 45 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.75 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 55 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | RCT reconstruction |
|---|---|
| CTF correction | Details: whole micrograph - unitilted images |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: Appion, Spider, Sparx, Eman2 / Number images used: 229 |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)