[English] 日本語
 Yorodumi
Yorodumi- EMDB-60063: Caenorhabditis elegans ACR-23 in betaine and monepantel bound state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Caenorhabditis elegans ACR-23 in betaine and monepantel bound state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHighly calcium permeable postsynaptic nicotinic acetylcholine receptors / Neurotransmitter receptors and postsynaptic signal transmission / excitatory extracellular ligand-gated monoatomic ion channel activity / transmembrane transporter complex / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / electron transport chain / transmembrane signaling receptor activity / monoatomic ion transmembrane transport / chemical synaptic transmission ...Highly calcium permeable postsynaptic nicotinic acetylcholine receptors / Neurotransmitter receptors and postsynaptic signal transmission / excitatory extracellular ligand-gated monoatomic ion channel activity / transmembrane transporter complex / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / electron transport chain / transmembrane signaling receptor activity / monoatomic ion transmembrane transport / chemical synaptic transmission / electron transfer activity / periplasmic space / neuron projection / postsynapse / iron ion binding / heme binding / synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Chen QF / Liu FL / Li TY / Gong HH / Guo F / Liu S | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structural insights into the molecular effects of the anthelmintics monepantel and betaine on the Caenorhabditis elegans acetylcholine receptor ACR-23. Authors: Fenglian Liu / Tianyu Li / Huihui Gong / Fei Tian / Yan Bai / Haowei Wang / Chonglin Yang / Yang Li / Fei Guo / Sheng Liu / Qingfeng Chen /  Abstract: Anthelmintics are drugs used for controlling pathogenic helminths in animals and plants. The natural compound betaine and the recently developed synthetic compound monepantel are both anthelmintics ...Anthelmintics are drugs used for controlling pathogenic helminths in animals and plants. The natural compound betaine and the recently developed synthetic compound monepantel are both anthelmintics that target the acetylcholine receptor ACR-23 and its homologs in nematodes. Here, we present cryo-electron microscopy structures of ACR-23 in apo, betaine-bound, and betaine- and monepantel-bound states. We show that ACR-23 forms a homo-pentameric channel, similar to some other pentameric ligand-gated ion channels (pLGICs). While betaine molecules are bound to the classical neurotransmitter sites in the inter-subunit interfaces in the extracellular domain, monepantel molecules are bound to allosteric sites formed in the inter-subunit interfaces in the transmembrane domain of the receptor. Although the pore remains closed in betaine-bound state, monepantel binding results in an open channel by wedging into the cleft between the transmembrane domains of two neighboring subunits, which causes dilation of the ion conduction pore. By combining structural analyses with site-directed mutagenesis, electrophysiology and in vivo locomotion assays, we provide insights into the mechanism of action of the anthelmintics monepantel and betaine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60063.map.gz emd_60063.map.gz | 566.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60063-v30.xml emd-60063-v30.xml emd-60063.xml emd-60063.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60063.png emd_60063.png | 93.4 KB | ||
| Filedesc metadata |  emd-60063.cif.gz emd-60063.cif.gz | 7 KB | ||
| Others |  emd_60063_half_map_1.map.gz emd_60063_half_map_1.map.gz emd_60063_half_map_2.map.gz emd_60063_half_map_2.map.gz | 557 MB 557 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60063 http://ftp.pdbj.org/pub/emdb/structures/EMD-60063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60063 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60063 | HTTPS FTP |
-Related structure data
| Related structure data |  8zfkMC  8zflC  8zfmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60063.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60063.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
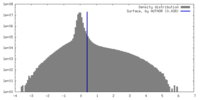
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60063_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
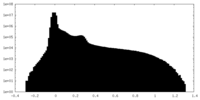
| Projections & Slices |
| ||||||||||||
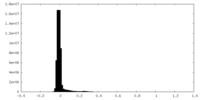
| Density Histograms |
-Half map: #1
| File | emd_60063_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
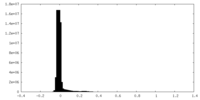
| Density Histograms |
- Sample components
Sample components
-Entire : acetylcholine receptor like-23(ACR-23)
| Entire | Name: acetylcholine receptor like-23(ACR-23) |
|---|---|
| Components |
|
-Supramolecule #1: acetylcholine receptor like-23(ACR-23)
| Supramolecule | Name: acetylcholine receptor like-23(ACR-23) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67 KDa |
-Macromolecule #1: Betaine receptor acr-23,Soluble cytochrome b562
| Macromolecule | Name: Betaine receptor acr-23,Soluble cytochrome b562 / type: protein_or_peptide / ID: 1 Details: Residues 387-500 correspond to the bril expression tag, whereas resides 604-609 correspond to the thrombin cleavage site, resides 599-603 and 610-612 correspond to flexible linkers, resides ...Details: Residues 387-500 correspond to the bril expression tag, whereas resides 604-609 correspond to the thrombin cleavage site, resides 599-603 and 610-612 correspond to flexible linkers, resides 613-620 correspond to the strep ll tag. Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 70.582281 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MHRIYTFLIF ISQLALGLSN NPDIPIQYEL ANNIMENYQK GLIPKVRKGS PINVTLSLQL YQIIQVNEPQ QYLLLNAWAV ERWVDQMLG WDPSEFDNET EIMARHDDIW LPDTTLYNSL EMDDSASKKL THVKLTTLGK NQGAMVELLY PTIYKISCLL N LKYFPFDT ...String: MHRIYTFLIF ISQLALGLSN NPDIPIQYEL ANNIMENYQK GLIPKVRKGS PINVTLSLQL YQIIQVNEPQ QYLLLNAWAV ERWVDQMLG WDPSEFDNET EIMARHDDIW LPDTTLYNSL EMDDSASKKL THVKLTTLGK NQGAMVELLY PTIYKISCLL N LKYFPFDT QTCRMTFGSW SFDNSLIDYF PRTFTNGPIG LANFLENDAW SVLGTKVNRE EKKYTCCPVN YTLLHYDVVI QR KPLYYVL NLIAPTAVIT FISIIGFFTS SSVHDLRQEK ITLGITTLLS MSIMIFMVSD KMPSTSTCVP LIALFYTLMI TII SVGTLA ASSVIFVQKL GSIGNPPASK TMKWTHRIAP FVLIQMPLVM KQAYAKRAKE EKHRKRMSRK MAEAGAMADL EDNW ETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDFR HGFDILVGQI DDALKLANEG KVKEA QAAA EQLKTTRNAY IQKYLSQTSE TFAAPMDTSF TESLHIPELN RVASSNSIQS VLKPTEIQLT PYCTRNIVEL EWDWVA AVL ERVFLIFFTI CFLFSAIGIN LYGWYIWYTE NHFLFLEGTK LVPRGSSSGW SHPQFEK UniProtKB: Betaine receptor acr-23, Soluble cytochrome b562, Betaine receptor acr-23 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: ~{N}-[(2~{S})-2-cyano-1-[5-cyano-2-(trifluoromethyl)phenoxy]propa...
| Macromolecule | Name: ~{N}-[(2~{S})-2-cyano-1-[5-cyano-2-(trifluoromethyl)phenoxy]propan-2-yl]-4-(trifluoromethylsulfanyl)benzamide type: ligand / ID: 4 / Number of copies: 5 / Formula: A1D8E |
|---|---|
| Molecular weight | Theoretical: 473.392 Da |
-Macromolecule #5: TRIMETHYL GLYCINE
| Macromolecule | Name: TRIMETHYL GLYCINE / type: ligand / ID: 5 / Number of copies: 5 / Formula: BET |
|---|---|
| Molecular weight | Theoretical: 118.154 Da |
| Chemical component information |  ChemComp-BET: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 15 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.00 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)