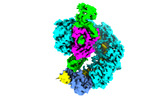
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Cas8-HNH system at target free state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | a protein complex / ANTIVIRAL PROTEIN | |||||||||
| Biological species |  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Zhang H / Zhu H / Li X / Liu Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structural basis for the type I-F Cas8-HNH system. Authors: Xuzichao Li / Yanan Liu / Jie Han / Lingling Zhang / Zhikun Liu / Lin Wang / Shuqin Zhang / Qian Zhang / Pengyu Fu / Hang Yin / Hongtao Zhu / Heng Zhang /  Abstract: The Cas3 nuclease is utilized by canonical type I CRISPR-Cas systems for processive target DNA degradation, while a newly identified type I-F CRISPR variant employs an HNH nuclease domain from the ...The Cas3 nuclease is utilized by canonical type I CRISPR-Cas systems for processive target DNA degradation, while a newly identified type I-F CRISPR variant employs an HNH nuclease domain from the natural fusion Cas8-HNH protein for precise target cleavage both in vitro and in human cells. Here, we report multiple cryo-electron microscopy structures of the type I-F Cas8-HNH system at different functional states. The Cas8-HNH Cascade complex adopts an overall G-shaped architecture, with the HNH domain occupying the C-terminal helical bundle domain (HB) of the Cas8 protein in canonical type I systems. The Linker region connecting Cas8-NTD and HNH domains adopts a rigid conformation and interacts with the Cas7.6 subunit, enabling the HNH domain to be in a functional position. The full R-loop formation displaces the HNH domain away from the Cas6 subunit, thus activating the target DNA cleavage. Importantly, our results demonstrate that precise target cleavage is dictated by a C-terminal helix of the HNH domain. Together, our work not only delineates the structural basis for target recognition and activation of the type I-F Cas8-HNH system, but also guides further developments leveraging this system for precise DNA editing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60017.map.gz emd_60017.map.gz | 155.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60017-v30.xml emd-60017-v30.xml emd-60017.xml emd-60017.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60017_fsc.xml emd_60017_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_60017.png emd_60017.png | 53.2 KB | ||
| Filedesc metadata |  emd-60017.cif.gz emd-60017.cif.gz | 6 KB | ||
| Others |  emd_60017_half_map_1.map.gz emd_60017_half_map_1.map.gz emd_60017_half_map_2.map.gz emd_60017_half_map_2.map.gz | 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60017 http://ftp.pdbj.org/pub/emdb/structures/EMD-60017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60017 | HTTPS FTP |
-Validation report
| Summary document |  emd_60017_validation.pdf.gz emd_60017_validation.pdf.gz | 861.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60017_full_validation.pdf.gz emd_60017_full_validation.pdf.gz | 861 KB | Display | |
| Data in XML |  emd_60017_validation.xml.gz emd_60017_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_60017_validation.cif.gz emd_60017_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60017 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60017 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60017 | HTTPS FTP |
-Related structure data
| Related structure data |  8zdyMC  8z0kC  8z0lC  8znrC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60017.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60017.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : a protein
| Entire | Name: a protein |
|---|---|
| Components |
|
-Supramolecule #1: a protein
| Supramolecule | Name: a protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
-Macromolecule #1: a protein
| Macromolecule | Name: a protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 38.700172 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAANKKATNV TLKSRPENLS FARCLNTTEA KFWQTDFLKR HTFKLPLLIT DKAVLASKGH EMPPDKLEKE IMDPNPQKSQ SCTLSTECD TLRIDFGIKV LPVKESMYSC SDYNYRTAIY QKIDEYIAED GFLTLAKRYV NNIANARFLW RNRKGAEIIE T IVTIEDKE ...String: MAANKKATNV TLKSRPENLS FARCLNTTEA KFWQTDFLKR HTFKLPLLIT DKAVLASKGH EMPPDKLEKE IMDPNPQKSQ SCTLSTECD TLRIDFGIKV LPVKESMYSC SDYNYRTAIY QKIDEYIAED GFLTLAKRYV NNIANARFLW RNRKGAEIIE T IVTIEDKE YPSFNSKSFN LDTFVEDNAT INEIAQQIAD TFAGKREYLN IYVTCFVKIG CAMEVYPSQE MTFDDDDKGK KL FKFEGSA GMHSQKINNA LRTIDTWYPD YTTYEFPIPV ENYGAARSIG IPFRPDTKSF YKLIDRMILK NEDLPIEDKH YVM AILIRG GMFSKKQEK |
-Macromolecule #2: a protein
| Macromolecule | Name: a protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 39.339742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRNKILAAI SQKIPEEQKI NKYIEGLFQS IDKNHLATHV AKFTETNSPG NIGAYDILSS DMNCGYLDTA NAGWKEPDIV TNDAKYKRP QGFVAMEMSD GRTVMEHLQE DSAELRHEME ELTDKYDEIR DGILNMPSMQ PYRTNQFIKQ VFFPVGGSYH L LSILPSTV ...String: MLRNKILAAI SQKIPEEQKI NKYIEGLFQS IDKNHLATHV AKFTETNSPG NIGAYDILSS DMNCGYLDTA NAGWKEPDIV TNDAKYKRP QGFVAMEMSD GRTVMEHLQE DSAELRHEME ELTDKYDEIR DGILNMPSMQ PYRTNQFIKQ VFFPVGGSYH L LSILPSTV LNYEVSDRLY RSKIPKIRLR LLSSNAASTT GSRLVSKNKW PLVFQALPPK FLEKNLAKAL DKEYLLPDIN ID ELEGVDN GCLIDEALLP LIIDEGKRKG EGNYRPRHLR DERKEETVQA FLDKYGYCNI PVGYEVHHIV PLSQGGADSI KNM IMLSIE HHERVTEAHA SYFKWRNT |
-Macromolecule #3: a protein
| Macromolecule | Name: a protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 28.703135 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMKGYILLEK VNIENANAFN NIIVGIPAIT SFLGFARALE RKLNAKEIAI RINGVGLEFH EYELKGYKNK RGQYVTSCPL PGSIPGQNE KKLDAHIMNQ AYIDLNMSFL LEVEGPHVDM STCKSIKSTM ETLRIAGGII RNYKKIRLID TLADIPYGYF L TLRQDNLN ...String: MMKGYILLEK VNIENANAFN NIIVGIPAIT SFLGFARALE RKLNAKEIAI RINGVGLEFH EYELKGYKNK RGQYVTSCPL PGSIPGQNE KKLDAHIMNQ AYIDLNMSFL LEVEGPHVDM STCKSIKSTM ETLRIAGGII RNYKKIRLID TLADIPYGYF L TLRQDNLN DAAGDDMLDK MIHALQQEDT LVPIAVGFKA LSEVGHVEGQ RDPEKDHCFV ESIFSLGGFE CSKILEDINS CL WRYKTEE GLYLCTII |
-Macromolecule #4: a protein
| Macromolecule | Name: a protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 20.735873 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFSQILIIKP GTGISPNIII SEDIFPVLHS LFVEHDKKFG ITFPAYSFDK KGHLGNIIEV LSEDKEALAS LCLEEHLAEV TDYVKVKKE ITFTDDYVLF KRIREENQYE TTARRMRKRG HTELGRPLEM HIKKKNQQIF CHAYIKVKSA STGQSYNIFL A PTDIKHGS FSAYGLLRGD THA |
-Macromolecule #5: RNA (58-MER)
| Macromolecule | Name: RNA (58-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Selenomonas sp. (bacteria) Selenomonas sp. (bacteria) |
| Molecular weight | Theoretical: 22.398293 KDa |
| Sequence | String: GUUUAGAAGG AUUGCCGUCA GGAAAUUAGG UGCGCUUAGC AGUGUACCGC CGGAUAGGCG GUUUAGAAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.2 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)