[English] 日本語
 Yorodumi
Yorodumi- EMDB-5772: A Two-Pronged Structural Analysis of Retroviral Maturation Indica... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5772 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A Two-Pronged Structural Analysis of Retroviral Maturation Indicates that Core Formation Proceeds by a Disassembly-Reassembly Pathway Rather than a Displacive Transition | |||||||||
 Map data Map data | T=1 icosahedral assembly of Rous sarcoma virus CA-SP protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / Rous Sarcoma Virus Structure / in vitro assembled capsids / spacer peptide | |||||||||
| Biological species |  Rous sarcoma virus Rous sarcoma virus | |||||||||
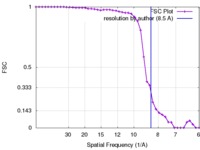
| Method | single particle reconstruction / cryo EM / Resolution: 8.5 Å | |||||||||
 Authors Authors | Keller PW / Huang RK / England M / Waki K / Cheng N / Heymann JB / Craven RC / Freed EO / Steven AC | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. Authors: Paul W Keller / Rick K Huang / Matthew R England / Kayoko Waki / Naiqian Cheng / J Bernard Heymann / Rebecca C Craven / Eric O Freed / Alasdair C Steven /  Abstract: Retrovirus maturation involves sequential cleavages of the Gag polyprotein, initially arrayed in a spherical shell, leading to formation of capsids with polyhedral or conical morphology. Evidence ...Retrovirus maturation involves sequential cleavages of the Gag polyprotein, initially arrayed in a spherical shell, leading to formation of capsids with polyhedral or conical morphology. Evidence suggests that capsids assemble de novo inside maturing virions from dissociated capsid (CA) protein, but the possibility persists of a displacive pathway in which the CA shell remains assembled but is remodeled. Inhibition of the final cleavage between CA and spacer peptide SP1/SP blocks the production of mature capsids. We investigated whether retention of SP might render CA assembly incompetent by testing the ability of Rous sarcoma virus (RSV) CA-SP to assemble in vitro into icosahedral capsids. Capsids were indeed assembled and were indistinguishable from those formed by CA alone, indicating that SP was disordered. We also used cryo-electron tomography to characterize HIV-1 particles produced in the presence of maturation inhibitor PF-46396 or with the cleavage-blocking CA5 mutation. Inhibitor-treated virions have a shell that resembles the CA layer of the immature Gag shell but is less complete. Some CA protein is generated but usually not enough for a mature core to assemble. We propose that inhibitors like PF-46396 bind to the Gag lattice where they deny the protease access to the CA-SP1 cleavage site and prevent the release of CA. CA5 particles, which exhibit no cleavage at the CA-SP1 site, have spheroidal shells with relatively thin walls. It appears that this lattice progresses displacively toward a mature-like state but produces neither conical cores nor infectious virions. These observations support the disassembly-reassembly pathway for core formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5772.map.gz emd_5772.map.gz | 55.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5772-v30.xml emd-5772-v30.xml emd-5772.xml emd-5772.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_5772_fsc.xml emd_5772_fsc.xml | 3.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_5772.jpg emd_5772.jpg | 58.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5772 http://ftp.pdbj.org/pub/emdb/structures/EMD-5772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5772 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5772.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5772.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T=1 icosahedral assembly of Rous sarcoma virus CA-SP protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : T=1 icosahedral assembly of Rous sarcoma virus capsid proteins wi...
| Entire | Name: T=1 icosahedral assembly of Rous sarcoma virus capsid proteins with spacer peptide |
|---|---|
| Components |
|
-Supramolecule #1000: T=1 icosahedral assembly of Rous sarcoma virus capsid proteins wi...
| Supramolecule | Name: T=1 icosahedral assembly of Rous sarcoma virus capsid proteins with spacer peptide type: sample / ID: 1000 Oligomeric state: T=1 icosahedral shell with 60 subunits forming 12 pentamers Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 1.51 MDa |
-Supramolecule #1: Rous sarcoma virus
| Supramolecule | Name: Rous sarcoma virus / type: virus / ID: 1 / Details: icosahedral assembly of CA-SP protein / NCBI-ID: 11886 / Sci species name: Rous sarcoma virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  |
| Molecular weight | Theoretical: 1.51 MDa |
| Virus shell | Shell ID: 1 / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 10 mM Tris-HCl, 75 mM sodium chloride, 0.05 mM EDTA, 0.5 M sodium phosphate |
| Grid | Details: Holey carbon film on R2/2 400 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 93.15 K / Instrument: LEICA KF80 / Details: Vitrification carried out in nitrogen atmosphere. Method: 4.0 microliter sample dropped onto grid, blotted on one side for 2 second, then plunged. |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 93.15 K |
| Date | Sep 24, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 17 / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | RSV CA NTD and CTD structures were rigid-body fitted into the T=1 density map using Chimera fit to map tools. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | RSV CA NTD and CTD structures were rigid-body fitted into the T=1 density map using Chimera fit to map tools. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)