[English] 日本語
 Yorodumi
Yorodumi- EMDB-5694: Type III secretion system (Injectisome) of Yersinia enterocolitic... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5694 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type III secretion system (Injectisome) of Yersinia enterocolitica in situ | |||||||||
 Map data Map data | In situ structure of the Yersinia enterocolitica Injectisome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | T3SS / YscC / YscD / YscJ / YscV / cryo electron tomography / sub-volume averaging. | |||||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Kudryashev M / Stenta M / Schmelz S / Amstutz M / Wiesand U / Castano-Diez D / Degiacomi M / Muennich S / Bleck CKE / Kowal J ...Kudryashev M / Stenta M / Schmelz S / Amstutz M / Wiesand U / Castano-Diez D / Degiacomi M / Muennich S / Bleck CKE / Kowal J / Diepold A / Heinz DW / Dal Peraro M / Cornelis GR / Stahlberg H | |||||||||
 Citation Citation |  Journal: Elife / Year: 2013 Journal: Elife / Year: 2013Title: In situ structural analysis of the Yersinia enterocolitica injectisome. Authors: Mikhail Kudryashev / Marco Stenta / Stefan Schmelz / Marlise Amstutz / Ulrich Wiesand / Daniel Castaño-Díez / Matteo T Degiacomi / Stefan Münnich / Christopher Ke Bleck / Julia Kowal / ...Authors: Mikhail Kudryashev / Marco Stenta / Stefan Schmelz / Marlise Amstutz / Ulrich Wiesand / Daniel Castaño-Díez / Matteo T Degiacomi / Stefan Münnich / Christopher Ke Bleck / Julia Kowal / Andreas Diepold / Dirk W Heinz / Matteo Dal Peraro / Guy R Cornelis / Henning Stahlberg /  Abstract: Injectisomes are multi-protein transmembrane machines allowing pathogenic bacteria to inject effector proteins into eukaryotic host cells, a process called type III secretion. Here we present the ...Injectisomes are multi-protein transmembrane machines allowing pathogenic bacteria to inject effector proteins into eukaryotic host cells, a process called type III secretion. Here we present the first three-dimensional structure of Yersinia enterocolitica and Shigella flexneri injectisomes in situ and the first structural analysis of the Yersinia injectisome. Unexpectedly, basal bodies of injectisomes inside the bacterial cells showed length variations of 20%. The in situ structures of the Y. enterocolitica and S. flexneri injectisomes had similar dimensions and were significantly longer than the isolated structures of related injectisomes. The crystal structure of the inner membrane injectisome component YscD appeared elongated compared to a homologous protein, and molecular dynamics simulations documented its elongation elasticity. The ring-shaped secretin YscC at the outer membrane was stretched by 30-40% in situ, compared to its isolated liposome-embedded conformation. We suggest that elasticity is critical for some two-membrane spanning protein complexes to cope with variations in the intermembrane distance. DOI:http://dx.doi.org/10.7554/eLife.00792.001. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5694.map.gz emd_5694.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5694-v30.xml emd-5694-v30.xml emd-5694.xml emd-5694.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5694_1.jpg emd_5694_1.jpg | 29.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5694 http://ftp.pdbj.org/pub/emdb/structures/EMD-5694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5694 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5694.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5694.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In situ structure of the Yersinia enterocolitica Injectisome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
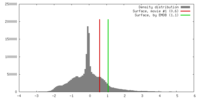
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yersinia enterocolitica intact bacterial cells containing type II...
| Entire | Name: Yersinia enterocolitica intact bacterial cells containing type III secretion injectisomes |
|---|---|
| Components |
|
-Supramolecule #1000: Yersinia enterocolitica intact bacterial cells containing type II...
| Supramolecule | Name: Yersinia enterocolitica intact bacterial cells containing type III secretion injectisomes type: sample / ID: 1000 Details: Bacteria were taken from cell culture and were rapidly prepared for cryo imaging Oligomeric state: various / Number unique components: 9 |
|---|
-Macromolecule #1: Bacterial Type III Secretion System
| Macromolecule | Name: Bacterial Type III Secretion System / type: protein_or_peptide / ID: 1 / Name.synonym: Injectisome, T3SS / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | Details: PBS |
|---|---|
| Grid | Details: Quantifoil holey carbon grids with various hole sizes |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 77 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Tridem / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Nov 1, 2009 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Number real images: 61 / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 19500 / Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.2 mm / Nominal defocus max: -15.0 µm / Nominal defocus min: -3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: OTHER / Number subtomograms used: 941 |
|---|---|
| CTF correction | Details: Each micrograph was phase flipped assuming the value of nominal defocus. Focal pair tomography was employed. |
| Final 3D classification | Number classes: 1 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)