[English] 日本語
 Yorodumi
Yorodumi- EMDB-5644: Two-dimensional crystals of sMgm1 over a monolayer with a lipid c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5644 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Two-dimensional crystals of sMgm1 over a monolayer with a lipid composition identical to that of the mitochondrial inner membrane | |||||||||
 Map data Map data | Three dimensional reconstruction from tilted images of a two-dimensional crystal of sMgm1 in the absence of substrate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondrial proteins / mitochondrial membrane fusion / mitochondrial cristae maintenance / yeast / DRP / sMgm1 | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial inner boundary membrane / Regulation of Apoptosis / mitochondrial outer membrane fusion / membrane bending / mitochondrion inheritance / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / GTPase-dependent fusogenic activity / heme transport ...mitochondrial inner boundary membrane / Regulation of Apoptosis / mitochondrial outer membrane fusion / membrane bending / mitochondrion inheritance / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / GTPase-dependent fusogenic activity / heme transport / dynamin GTPase / cristae formation / phosphatidic acid binding / cardiolipin binding / phosphatidylinositol-3,5-bisphosphate binding / mitochondrial fusion / mitochondrial membrane organization / phosphatidylserine binding / mitochondrial crista / mitochondrion organization / mitochondrial intermembrane space / microtubule binding / microtubule / mitochondrial outer membrane / mitochondrial inner membrane / GTPase activity / GTP binding / mitochondrion / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | electron crystallography / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Devay RM / Dominguez-Ramirez L / Lackner LL / Hoppins S / Stahlberg H / Nunnari J | |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2009 Journal: J Cell Biol / Year: 2009Title: Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. Authors: Rachel M DeVay / Lenin Dominguez-Ramirez / Laura L Lackner / Suzanne Hoppins / Henning Stahlberg / Jodi Nunnari /  Abstract: Two dynamin-related protein (DRP) families are essential for fusion of the outer and inner mitochondrial membranes, Fzo1 (yeast)/Mfn1/Mfn2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), respectively. ...Two dynamin-related protein (DRP) families are essential for fusion of the outer and inner mitochondrial membranes, Fzo1 (yeast)/Mfn1/Mfn2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), respectively. Fzo1/Mfns possess two medial transmembrane domains, which place their critical GTPase and coiled-coil domains in the cytosol. In contrast, Mgm1/Opa1 are present in cells as long (l) isoforms that are anchored via the N terminus to the inner membrane, and short (s) isoforms were predicted to be soluble in the intermembrane space. We addressed the roles of Mgm1 isoforms and how DRPs function in membrane fusion. Our analysis indicates that in the absence of a membrane, l- and s-Mgm1 both exist as inactive GTPase monomers, but that together in trans they form a functional dimer in a cardiolipin-dependent manner that is the building block for higher-order assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5644.map.gz emd_5644.map.gz | 56.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5644-v30.xml emd-5644-v30.xml emd-5644.xml emd-5644.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5644_1.jpg emd_5644_1.jpg | 64.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5644 http://ftp.pdbj.org/pub/emdb/structures/EMD-5644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5644 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5644 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5644.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5644.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three dimensional reconstruction from tilted images of a two-dimensional crystal of sMgm1 in the absence of substrate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
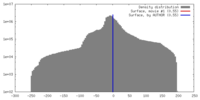
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 143 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Small isoform of yeast Mgm1
| Entire | Name: Small isoform of yeast Mgm1 |
|---|---|
| Components |
|
-Supramolecule #1000: Small isoform of yeast Mgm1
| Supramolecule | Name: Small isoform of yeast Mgm1 / type: sample / ID: 1000 Details: The sample aggregates as the oligomer observed by electron microscopy only in the presence of lipids, particularly those making up the inner mitochondrial membrane. Oligomeric state: trimer of dimers / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 76 MDa / Theoretical: 76 MDa / Method: Sedimentation |
-Macromolecule #1: Mitochondrial genome maintenance 1
| Macromolecule | Name: Mitochondrial genome maintenance 1 / type: protein_or_peptide / ID: 1 / Name.synonym: Dynamin-like GTPase MGM1, mitochondrial Details: Lipid monolayer with same composition as inner mitochondrial membranes Number of copies: 6 / Oligomeric state: Trimer of dimers / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 76 MDa / Theoretical: 76 MDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Dynamin-like GTPase MGM1, mitochondrial / GO: mitochondrial fusion InterPro: Dynamin, Dynamin, GTPase domain, Dynamin, GTPase region, conserved site, GTPase effector domain |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl |
| Staining | Type: NEGATIVE Details: Grids were floated on 2% uranyl acetate for 10 seconds twice. |
| Grid | Details: 200 mesh copper grid with thin carbon support, non-glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
| Details | Crystals were grown on a lipid monolayer. |
| Crystal formation | Details: Crystals were grown on a lipid monolayer. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Temperature | Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification. |
| Date | Jul 2, 2009 |
| Image recording | Number real images: 35 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: JEOL / Tilt angle max: 45 / Tilt series - Axis1 - Min angle: 0 ° / Tilt series - Axis1 - Max angle: 45 ° |
- Image processing
Image processing
| Details | Images were processed entirely using 2dx. |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Software - Name: 2DX |
| Crystal parameters | Unit cell - A: 200 Å / Unit cell - B: 200 Å / Unit cell - γ: 120 ° / Plane group: P 3 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)