[English] 日本語
 Yorodumi
Yorodumi- EMDB-5386: tmRNA translocation and MLD-loading on the ribosome: a 70S-tmRNA-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5386 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | tmRNA translocation and MLD-loading on the ribosome: a 70S-tmRNA-EF-G complex | |||||||||
 Map data Map data | Overall view of the 70S-tmRNA-SmpB-EF-G complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | trans-translation / MLD-loading / translocation / tmRNA / EF-G / SmpB | |||||||||
| Function / homology |  Function and homology information Function and homology informationstringent response / transcription antitermination factor activity, RNA binding / ornithine decarboxylase inhibitor activity / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity ...stringent response / transcription antitermination factor activity, RNA binding / ornithine decarboxylase inhibitor activity / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / translation repressor activity / negative regulation of translational initiation / regulation of mRNA stability / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / response to reactive oxygen species / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / DNA endonuclease activity / regulation of cell growth / DNA-templated transcription termination / response to radiation / maintenance of translational fidelity / mRNA 5'-UTR binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosome binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / hydrolase activity / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 8.3 Å | |||||||||
 Authors Authors | Ramrath DJF / Yamamoto H / Rother K / Wittek D / Pech M / Mielke T / Loerke J / Scheerer P / Ivanov P / Teraoka Y ...Ramrath DJF / Yamamoto H / Rother K / Wittek D / Pech M / Mielke T / Loerke J / Scheerer P / Ivanov P / Teraoka Y / Shpanchenko O / Nierhaus KH / Spahn CMT | |||||||||
 Citation Citation |  Journal: Nature / Year: 2012 Journal: Nature / Year: 2012Title: The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Authors: David J F Ramrath / Hiroshi Yamamoto / Kristian Rother / Daniela Wittek / Markus Pech / Thorsten Mielke / Justus Loerke / Patrick Scheerer / Pavel Ivanov / Yoshika Teraoka / Olga Shpanchenko ...Authors: David J F Ramrath / Hiroshi Yamamoto / Kristian Rother / Daniela Wittek / Markus Pech / Thorsten Mielke / Justus Loerke / Patrick Scheerer / Pavel Ivanov / Yoshika Teraoka / Olga Shpanchenko / Knud H Nierhaus / Christian M T Spahn /  Abstract: Bacterial ribosomes stalled at the 3' end of malfunctioning messenger RNAs can be rescued by transfer-messenger RNA (tmRNA)-mediated trans-translation. The SmpB protein forms a complex with the ...Bacterial ribosomes stalled at the 3' end of malfunctioning messenger RNAs can be rescued by transfer-messenger RNA (tmRNA)-mediated trans-translation. The SmpB protein forms a complex with the tmRNA, and the transfer-RNA-like domain (TLD) of the tmRNA then enters the A site of the ribosome. Subsequently, the TLD-SmpB module is translocated to the P site, a process that is facilitated by the elongation factor EF-G, and translation is switched to the mRNA-like domain (MLD) of the tmRNA. Accurate loading of the MLD into the mRNA path is an unusual initiation mechanism. Despite various snapshots of different ribosome-tmRNA complexes at low to intermediate resolution, it is unclear how the large, highly structured tmRNA is translocated and how the MLD is loaded. Here we present a cryo-electron microscopy reconstruction of a fusidic-acid-stalled ribosomal 70S-tmRNA-SmpB-EF-G complex (carrying both of the large ligands, that is, EF-G and tmRNA) at 8.3 Å resolution. This post-translocational intermediate (TI(POST)) presents the TLD-SmpB module in an intrasubunit ap/P hybrid site and a tRNA(fMet) in an intrasubunit pe/E hybrid site. Conformational changes in the ribosome and tmRNA occur in the intersubunit space and on the solvent side. The key underlying event is a unique extra-large swivel movement of the 30S head, which is crucial for both tmRNA-SmpB translocation and MLD loading, thereby coupling translocation to MLD loading. This mechanism exemplifies the versatile, dynamic nature of the ribosome, and it shows that the conformational modes of the ribosome that normally drive canonical translation can also be used in a modified form to facilitate more complex tasks in specialized non-canonical pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5386.map.gz emd_5386.map.gz | 80.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5386-v30.xml emd-5386-v30.xml emd-5386.xml emd-5386.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5386_1.png emd_5386_1.png | 634.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5386 http://ftp.pdbj.org/pub/emdb/structures/EMD-5386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5386 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5386 | HTTPS FTP |
-Related structure data
| Related structure data |  4v6tMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5386.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5386.map.gz / Format: CCP4 / Size: 100.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Overall view of the 70S-tmRNA-SmpB-EF-G complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
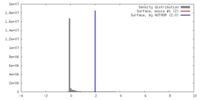
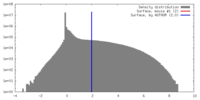
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of 70S-tmRNA-SmpB-EF-G
| Entire | Name: Complex of 70S-tmRNA-SmpB-EF-G |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of 70S-tmRNA-SmpB-EF-G
| Supramolecule | Name: Complex of 70S-tmRNA-SmpB-EF-G / type: sample / ID: 1000 / Details: the sample was flash frozen in liquid ethane / Oligomeric state: Monomer / Number unique components: 5 |
|---|---|
| Molecular weight | Experimental: 3 MDa / Theoretical: 3 MDa / Method: Sedimentation |
-Supramolecule #1: Ribosome
| Supramolecule | Name: Ribosome / type: complex / ID: 1 / Name.synonym: 70S-tmRNA-EFG / Details: the complex was stalled with fusidic acid / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 3 MDa / Theoretical: 3.05 MDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL |
|---|---|
| Buffer | pH: 7.6 Details: 20 mM HEPES-KOH, pH 7.6, 4.5 mM Mg(Ac)2, 150 mM NH4Ac, 4 mM b-mercaptoethanol, 2 mM spermidine, and 0.05 mM spermine |
| Staining | Type: NEGATIVE / Details: flash frozen in liquid ethane |
| Grid | Details: Quantifoil R3-3 Cu 300 mesh with 2 nm carbon support film |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 96.15 K / Instrument: OTHER / Details: Vitrification instrument: FEI VITROBOT / Method: blot for 10 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 77.15 K / Max: 81.15 K / Average: 77.15 K |
| Date | Dec 8, 2009 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 4.7 µm / Number real images: 302 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 39000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Cartridge / Specimen holder model: GATAN HELIUM |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Defocus groups |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.3 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider / Number images used: 68842 |
-Atomic model buiding 1
| Initial model | PDB ID:  3r8s |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid Body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross-correlation |
| Output model |  PDB-4v6t: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)