+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of rat SLCO2A1 bound to dinoprost. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | OATP2A1 / plasma membrane localized / prostaglandin transporter / PGT / major facilitator superfamily. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationOrganic anion transport by SLCO transporters / sodium-independent organic anion transport / : / prostaglandin transport / prostaglandin transmembrane transporter activity / basal plasma membrane / basolateral plasma membrane / lysosome Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Deme JC / Joshi C / Lea SM / Newstead S | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for prostaglandin and drug transport via SLCO2A1. Authors: Joshi C / Deme JC / Nakamura Y / Hsu W-T / Goult J / Kato T / Parker JL / Biggin PC / Lea SM / Nakanishi T / Newstead S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53485.map.gz emd_53485.map.gz | 194.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53485-v30.xml emd-53485-v30.xml emd-53485.xml emd-53485.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53485_fsc.xml emd_53485_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_53485.png emd_53485.png | 80.8 KB | ||
| Filedesc metadata |  emd-53485.cif.gz emd-53485.cif.gz | 6.6 KB | ||
| Others |  emd_53485_additional_1.map.gz emd_53485_additional_1.map.gz emd_53485_half_map_1.map.gz emd_53485_half_map_1.map.gz emd_53485_half_map_2.map.gz emd_53485_half_map_2.map.gz | 217 MB 213.2 MB 213.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53485 http://ftp.pdbj.org/pub/emdb/structures/EMD-53485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53485 | HTTPS FTP |
-Related structure data
| Related structure data |  9r0nMC  9qzoC  9r07C  9r0aC  9r0iC  9r0mC  9r0oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_53485.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53485.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.732 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_53485_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_53485_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_53485_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SLCO2A1 protein in detergent-lipid micelle bound to prostaglandin...
| Entire | Name: SLCO2A1 protein in detergent-lipid micelle bound to prostaglandin F2alpha, dinoprost. |
|---|---|
| Components |
|
-Supramolecule #1: SLCO2A1 protein in detergent-lipid micelle bound to prostaglandin...
| Supramolecule | Name: SLCO2A1 protein in detergent-lipid micelle bound to prostaglandin F2alpha, dinoprost. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier organic anion transporter family member 2A1
| Macromolecule | Name: Solute carrier organic anion transporter family member 2A1 type: protein_or_peptide / ID: 1 Details: Fusion protein: Avi tag (GLNDIFEAQKIEWHE) Linker as restriction site used for cloning (XhoI): 3'-SLCO2A1-Avitag-LEGG-5' Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 72.615117 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGLLLKPGAR QGSGTSSVPD RRCPRSVFSN IKVFVLCHGL LQLCQLLYSA YFKSSLTTIE KRFGLSSSSS GLISSLNEIS NATLIIFIS YFGSRVNRPR MIGIGGLLLA AGAFVLTLPH FLSEPYQYTS TTDGNRSSFQ TDLCQKHFGA LPPSKCHSTV P DTHKETSS ...String: MGLLLKPGAR QGSGTSSVPD RRCPRSVFSN IKVFVLCHGL LQLCQLLYSA YFKSSLTTIE KRFGLSSSSS GLISSLNEIS NATLIIFIS YFGSRVNRPR MIGIGGLLLA AGAFVLTLPH FLSEPYQYTS TTDGNRSSFQ TDLCQKHFGA LPPSKCHSTV P DTHKETSS LWGLMVVAQL LAGIGTVPIQ PFGISYVDDF AEPTNSPLYI SILFAIAVFG PAFGYLLGSV MLRIFVDYGR VD TATVNLS PGDPRWIGAW WLGLLISSGF LIVTSLPFFF FPRAMSRGAE RSVTAEETMQ TEEDKSRGSL MDFIKRFPRI FLR LLMNPL FMLVVLSQCT FSSVIAGLST FLNKFLEKQY GATAAYANFL IGAVNLPAAA LGMLFGGILM KRFVFPLQTI PRVA ATIIT ISMILCVPLF FMGCSTSAVA EVYPPSTSSS IHPQQPPACR RDCSCPDSFF HPVCGDNGVE YVSPCHAGCS STNTS SEAS KEPIYLNCSC VSGGSASAKT GSCPTSCAQL LLPSIFLISF AALIACISHN PLYMMVLRVV NQDEKSFAIG VQFLLM RLL AWLPAPSLYG LLIDSSCVRW NYLCSGRRGA CAYYDNDALR NRYLGLQMVY KALGTLLLFF ISWRMKKNRE YSLQENT SG LIGLNDIFEA QKIEWHELEG G UniProtKB: Solute carrier organic anion transporter family member 2A1 |
-Macromolecule #2: (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-...
| Macromolecule | Name: (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid type: ligand / ID: 2 / Number of copies: 1 / Formula: UGU |
|---|---|
| Molecular weight | Theoretical: 354.481 Da |
| Chemical component information | 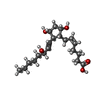 ChemComp-UGU: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 61.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-9r0n: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































