[English] 日本語
 Yorodumi
Yorodumi- EMDB-53384: Cryo-EM structure of the human NHA2-Fab complex bound to phloretin -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human NHA2-Fab complex bound to phloretin | |||||||||
 Map data Map data | NHA2_phloretin_J103 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Na+/H+ exchanger / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sodium ion homeostasis / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / flagellated sperm motility / : / sodium ion transport ...lithium:proton antiporter activity / lithium ion transport / positive regulation of osteoclast development / sodium ion homeostasis / sodium:proton antiporter activity / regulation of insulin secretion involved in cellular response to glucose stimulus / clathrin-dependent endocytosis / flagellated sperm motility / : / sodium ion transport / sperm principal piece / recycling endosome / mitochondrial membrane / Stimuli-sensing channels / recycling endosome membrane / synaptic vesicle membrane / monoatomic ion transmembrane transport / basolateral plasma membrane / apical plasma membrane / mitochondrial inner membrane / mitochondrion / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Jung S / Drew D | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2025 Journal: Int J Mol Sci / Year: 2025Title: Structure and Inhibition of the Human Na/H Exchanger SLC9B2. Authors: Sukkyeong Jung / Surabhi Kokane / Hang Li / So Iwata / Norimichi Nomura / David Drew /   Abstract: The sodium/proton exchanger NHA2, also known as SLC9B2, is important for insulin secretion, renal blood pressure regulation, and electrolyte retention. Recent structures of bison NHA2 has revealed ...The sodium/proton exchanger NHA2, also known as SLC9B2, is important for insulin secretion, renal blood pressure regulation, and electrolyte retention. Recent structures of bison NHA2 has revealed its unique 14-transmembrane helix architecture, which is different from SLC9A/NHE members made up from 13-TM helices. Sodium/proton exchangers are functional homodimers, and the additional N-terminal helix in NHA2 was found to alter homodimer assembly. Here, we present the cryo-electron microscopy structures of apo human NHA2 in complex with a Fab fragment and also with the inhibitor phloretin bound at 2.8 and 2.9 Å resolution, respectively. We show how phosphatidic acid (PA) lipids bind to the homodimer interface of NHA2 on the extracellular side, which we propose has a regulatory role linked to cell volume regulation. The ion binding site of human NHA2 has a salt bridge interaction between the ion binding aspartate D278 and R432, an interaction previously broken in the bison NHA2 structure, and these differences suggest a possible ion coupling mechanism. Lastly, the human NHA2 structure in complex with phloretin offers a template for structure-guided drug design, potentially leading to the development of more selective and potent NHA2 inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53384.map.gz emd_53384.map.gz | 120.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53384-v30.xml emd-53384-v30.xml emd-53384.xml emd-53384.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53384_fsc.xml emd_53384_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_53384.png emd_53384.png | 65.1 KB | ||
| Filedesc metadata |  emd-53384.cif.gz emd-53384.cif.gz | 6.6 KB | ||
| Others |  emd_53384_half_map_1.map.gz emd_53384_half_map_1.map.gz emd_53384_half_map_2.map.gz emd_53384_half_map_2.map.gz | 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53384 http://ftp.pdbj.org/pub/emdb/structures/EMD-53384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53384 | HTTPS FTP |
-Related structure data
| Related structure data |  9quwMC  9qubC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_53384.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53384.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NHA2_phloretin_J103 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A NHA2 phloretin J103
| File | emd_53384_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_A_NHA2_phloretin_J103 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B NHA2 phloretin J103
| File | emd_53384_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_B_NHA2_phloretin_J103 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : phloretin-bound complex of human NHA2 with Fab
| Entire | Name: phloretin-bound complex of human NHA2 with Fab |
|---|---|
| Components |
|
-Supramolecule #1: phloretin-bound complex of human NHA2 with Fab
| Supramolecule | Name: phloretin-bound complex of human NHA2 with Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fab light chain
| Macromolecule | Name: Fab light chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.715109 KDa |
| Sequence | String: DIVMTQTTSS LSASLGDRVT ISCRASQDIS NYLNWFQQKP DGTVKLLICY TSRLHSGVPS RFSGSGSGTD YSLTISNLEQ EDIATYFCQ QDSKHPWTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVMTQTTSS LSASLGDRVT ISCRASQDIS NYLNWFQQKP DGTVKLLICY TSRLHSGVPS RFSGSGSGTD YSLTISNLEQ EDIATYFCQ QDSKHPWTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Macromolecule #2: Fab heavy chain
| Macromolecule | Name: Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.757529 KDa |
| Sequence | String: EVQLQESGPE LVKPGASVKM SCKASGYTFT NYFIHWVKQK PGQGLEWIGY INPYNDITKF NEKFKGKATL TSDKSSRTAY MELSSLTSE DSAVYYCARC DGYYRYYAMD YWGQGTSVTV SSAKTTAPSV YPLAPVCGDT SGSSVTLGCL VKGYFPEPVT L TWNSGSLS ...String: EVQLQESGPE LVKPGASVKM SCKASGYTFT NYFIHWVKQK PGQGLEWIGY INPYNDITKF NEKFKGKATL TSDKSSRTAY MELSSLTSE DSAVYYCARC DGYYRYYAMD YWGQGTSVTV SSAKTTAPSV YPLAPVCGDT SGSSVTLGCL VKGYFPEPVT L TWNSGSLS SGVHTFPAVL QSDLYTLSSS VTVTSSTWPS QSITCNVAHP ASSTKVDKKI EPRGPTIKPC PPCKCPAPNL LG GPSVFIF PPKIKDVLM |
-Macromolecule #3: Sodium/hydrogen exchanger 9B2
| Macromolecule | Name: Sodium/hydrogen exchanger 9B2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.611672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDEDKRITY EDSEPSTGMN YTPSMHQEAQ EETVMKLKGI DANEPTEGSI LLKSSEKKLQ ETPTEANHVQ RLRQMLACPP HGLLDRVIT NVTIIVLLWA VVWSITGSEC LPGGNLFGII ILFYCAIIGG KLLGLIKLPT LPPLPSLLGM LLAGFLIRNI P VINDNVQI ...String: MGDEDKRITY EDSEPSTGMN YTPSMHQEAQ EETVMKLKGI DANEPTEGSI LLKSSEKKLQ ETPTEANHVQ RLRQMLACPP HGLLDRVIT NVTIIVLLWA VVWSITGSEC LPGGNLFGII ILFYCAIIGG KLLGLIKLPT LPPLPSLLGM LLAGFLIRNI P VINDNVQI KHKWSSSLRS IALSIILVRA GLGLDSKALK KLKGVCVRLS MGPCIVEACT SALLAHYLLG LPWQWGFILG FV LGAVSPA VVVPSMLLLQ GGGYGVEKGV PTLLMAAGSF DDILAITGFN TCLGIAFSTG STVFNVLRGV LEVVIGVATG SVL GFFIQY FPSRDQDKLV CKRTFLVLGL SVLAVFSSVH FGFPGSGGLC TLVMAFLAGM GWTSEKAEVE KIIAVAWDIF QPLL FGLIG AEVSIASLRP ETVGLCVATV GIAVLIRILT TFLMVCFAGF NLKEKIFISF AWLPKATVQA AIGSVALDTA RSHGE KQLE DYGMDVLTVA FLSILITAPI GSLLIGLLGP RLLQKVEHQN KDEEVQGETS VQV UniProtKB: Sodium/hydrogen exchanger 9B2 |
-Macromolecule #4: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE
| Macromolecule | Name: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE type: ligand / ID: 4 / Number of copies: 2 / Formula: LPP |
|---|---|
| Molecular weight | Theoretical: 648.891 Da |
| Chemical component information |  ChemComp-LPP: |
-Macromolecule #5: 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE
| Macromolecule | Name: 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE type: ligand / ID: 5 / Number of copies: 2 / Formula: G50 |
|---|---|
| Molecular weight | Theoretical: 274.269 Da |
| Chemical component information | 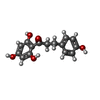 ChemComp-G50: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 65.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































