[English] 日本語
 Yorodumi
Yorodumi- EMDB-53371: Cryo-EM structure of the inward-open choline-bound state of choli... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the inward-open choline-bound state of choline/ethanolamine transporter FLVCR2 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Choline transporter / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationheme export / ethanolamine transmembrane transporter activity / choline transmembrane transporter activity / heme transmembrane transporter activity / choline transport / IgG binding / transport across blood-brain barrier / mitochondrial membrane / heme binding / endoplasmic reticulum membrane ...heme export / ethanolamine transmembrane transporter activity / choline transmembrane transporter activity / heme transmembrane transporter activity / choline transport / IgG binding / transport across blood-brain barrier / mitochondrial membrane / heme binding / endoplasmic reticulum membrane / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.39 Å | |||||||||||||||
 Authors Authors | Driller JH / Nel L / Pedersen BP | |||||||||||||||
| Funding support |  Denmark, European Union, 4 items Denmark, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Comparative and structural analysis of choline transport between eukaryotic choline transport protein families FLVCR and CTL Authors: Nel L / Driller JH / Driller R / Frain KM / Pedersen BP | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53371.map.gz emd_53371.map.gz | 25.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53371-v30.xml emd-53371-v30.xml emd-53371.xml emd-53371.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53371_fsc.xml emd_53371_fsc.xml | 6.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_53371.png emd_53371.png | 53.7 KB | ||
| Filedesc metadata |  emd-53371.cif.gz emd-53371.cif.gz | 6.1 KB | ||
| Others |  emd_53371_half_map_1.map.gz emd_53371_half_map_1.map.gz emd_53371_half_map_2.map.gz emd_53371_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53371 http://ftp.pdbj.org/pub/emdb/structures/EMD-53371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53371 | HTTPS FTP |
-Related structure data
| Related structure data |  9qu4MC  9qu3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_53371.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53371.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.294 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_53371_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_53371_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Choline/ethanolamine transporter FLVCR2 in DDM
| Entire | Name: Choline/ethanolamine transporter FLVCR2 in DDM |
|---|---|
| Components |
|
-Supramolecule #1: Choline/ethanolamine transporter FLVCR2 in DDM
| Supramolecule | Name: Choline/ethanolamine transporter FLVCR2 in DDM / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Immunoglobulin G-binding protein A,Choline/ethanolamine transport...
| Macromolecule | Name: Immunoglobulin G-binding protein A,Choline/ethanolamine transporter FLVCR2 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.364492 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGPMVNENKE QQNAFYEILS LPNLNEEQRN AFIQSLKDDP SQSANLLAEA KKLNEQQAAF YEILHLPNLN EEQRNAFIQS LKDDPSQSA NLLAEAKKLN EQQAAFYEIL HLPNLNEEQR NAFIQSLKDD PSQSANLLAE AKKLNELQKR RWAVVLVFSC Y SMCNSFQW ...String: MGPMVNENKE QQNAFYEILS LPNLNEEQRN AFIQSLKDDP SQSANLLAEA KKLNEQQAAF YEILHLPNLN EEQRNAFIQS LKDDPSQSA NLLAEAKKLN EQQAAFYEIL HLPNLNEEQR NAFIQSLKDD PSQSANLLAE AKKLNELQKR RWAVVLVFSC Y SMCNSFQW IQYGSINNIF MHFYGVSAFA IDWLSMCYML TYIPLLLPVA WLLEKFGLRT IALTGSALNC LGAWVKLGSL KP HLFPVTV VGQLICSVAQ VFILGMPSRI ASVWFGANEV STACSVAVFG NQLGIAIGFL VPPVLVPNIE DRDELAYHIS IMF YIIGGV ATLLLILVII VFKEKPKYPP SRAQSLSYAL TSPDASYLGS IARLFKNLNF VLLVITYGLN AGAFYALSTL LNRM VIWHY PGEEVNAGRI GLTIVIAGML GAVISGIWLD RSKTYKETTL VVYIMTLVGM VVYTFTLNLG HLWVVFITAG TMGFF MTGY LPLGFEFAVE LTYPESEGIS SGLLNISAQV FGIIFTISQG QIIDNYGTKP GNIFLCVFLT LGAALTAFIK ADLRRQ KAN KETLENKLQE EEEESNTSKV PTAVSEDHLG LVPR UniProtKB: Immunoglobulin G-binding protein A, Choline/ethanolamine transporter FLVCR2 |
-Macromolecule #2: CHOLINE ION
| Macromolecule | Name: CHOLINE ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CHT |
|---|---|
| Molecular weight | Theoretical: 104.171 Da |
| Chemical component information | 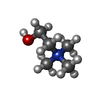 ChemComp-CHT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































