+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5290 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM map of DegQ 12-mer | |||||||||
 Map data Map data | This is a cryo-EM map of DegQ 12-mer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HtrA / 12-mer | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | |||||||||
 Authors Authors | Bai XC / Wang XJ / Ye YY / Pan XJ / Chang LF / Leng D / Sui SF | |||||||||
 Citation Citation |  Journal: Structure / Year: 2011 Journal: Structure / Year: 2011Title: Characterization of the structure and function of Escherichia coli DegQ as a representative of the DegQ-like proteases of bacterial HtrA family proteins. Authors: Xiao-chen Bai / Xi-jiang Pan / Xiao-jing Wang / Yun-ying Ye / Lei-fu Chang / Dong Leng / Jianlin Lei / Sen-Fang Sui /  Abstract: HtrA family proteins play a central role in protein quality control in the bacterial periplasmic space. DegQ-like proteases, a group of bacterial HtrA proteins, are characterized by a short LA loop ...HtrA family proteins play a central role in protein quality control in the bacterial periplasmic space. DegQ-like proteases, a group of bacterial HtrA proteins, are characterized by a short LA loop as compared with DegP-like proteases, and are found in many bacterial species. As a representative of the DegQ-like proteases, we report that Escherichia coli DegQ exists in vivo primarily as a trimer (substrate-free) or dodecamer (substrate-containing). Biochemical analysis of DegQ dodecamers revealed that the major copurified protein substrate is OmpA. Importantly, wild-type DegQ exhibited a much lower proteolytic activity, and thus higher chaperone-like activity, than DegP. Furthermore, using cryo-electron microscopy we determined high-resolution structures of DegQ 12- and 24-mers in the presence of substrate, thus revealing the structural mechanism by which DegQ moderates its proteolytic activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5290.map.gz emd_5290.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5290-v30.xml emd-5290-v30.xml emd-5290.xml emd-5290.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5290_1.jpg emd_5290_1.jpg | 219.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5290 http://ftp.pdbj.org/pub/emdb/structures/EMD-5290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5290 | HTTPS FTP |
-Validation report
| Summary document |  emd_5290_validation.pdf.gz emd_5290_validation.pdf.gz | 79.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5290_full_validation.pdf.gz emd_5290_full_validation.pdf.gz | 78.5 KB | Display | |
| Data in XML |  emd_5290_validation.xml.gz emd_5290_validation.xml.gz | 491 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5290 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5290 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5290 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5290 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5290.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5290.map.gz / Format: CCP4 / Size: 11.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a cryo-EM map of DegQ 12-mer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
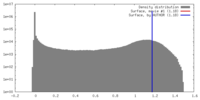
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DegQ
| Entire | Name: DegQ |
|---|---|
| Components |
|
-Supramolecule #1000: DegQ
| Supramolecule | Name: DegQ / type: sample / ID: 1000 / Oligomeric state: 12-mer / Number unique components: 12 |
|---|---|
| Molecular weight | Experimental: 500 KDa / Theoretical: 500 KDa / Method: size-exclusion chromatography |
-Macromolecule #1: DegQ
| Macromolecule | Name: DegQ / type: protein_or_peptide / ID: 1 / Name.synonym: DegQ / Number of copies: 12 / Oligomeric state: 12-mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 45 MDa / Theoretical: 45 MDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 40 mM Na2HPO4, 10 mM NaH2PO4, 25 mM NaCl |
| Grid | Details: 400 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Details: Vitrification instrument: Vitrobot Mark IV / Method: Blot for 1.5 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Jan 3, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 15 µm / Number real images: 315 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 102740 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.2 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 24604 |
| Final two d classification | Number classes: 180 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)