+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5269 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D reconstruction of yeast coatomer | |||||||||
 Map data Map data | 3D reconstruction of yeast coatomer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | coatomer / COP1 vesicle / vesicular trafficking / coat complex | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 36.0 Å | |||||||||
 Authors Authors | Yip CK / Walz T | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2011 Journal: J Mol Biol / Year: 2011Title: Molecular structure and flexibility of the yeast coatomer as revealed by electron microscopy. Authors: Calvin K Yip / Thomas Walz /  Abstract: Coat protein complex I (COPI)-coated vesicles, one of three major types of vesicular carriers in the cell, mediate the early secretory pathway and retrograde transport from the Golgi to the ...Coat protein complex I (COPI)-coated vesicles, one of three major types of vesicular carriers in the cell, mediate the early secretory pathway and retrograde transport from the Golgi to the endoplasmic reticulum. COPI vesicles are generated through activation of the regulatory GTPase Arf1 at the donor membrane and the subsequent recruitment of coatomer, a coat protein complex consisting of seven stably associated components. Coatomer functions in binding and sequestering cargo molecules and assembles into a polymeric protein shell that encompasses the surface of COPI vesicles. Little is known about the structural properties of this heptameric complex. We have isolated native yeast coatomer and examined its structure and subunit organization by single-particle electron microscopy. Our analyses provide the first three-dimensional picture of the complete coatomer and reveal substantial conformational flexibility likely to be critical for its scaffolding function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5269.map.gz emd_5269.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5269-v30.xml emd-5269-v30.xml emd-5269.xml emd-5269.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5269_1.png emd_5269_1.png | 25.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5269 http://ftp.pdbj.org/pub/emdb/structures/EMD-5269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5269 | HTTPS FTP |
-Validation report
| Summary document |  emd_5269_validation.pdf.gz emd_5269_validation.pdf.gz | 77.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5269_full_validation.pdf.gz emd_5269_full_validation.pdf.gz | 76.9 KB | Display | |
| Data in XML |  emd_5269_validation.xml.gz emd_5269_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5269 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5269 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5269.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5269.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of yeast coatomer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
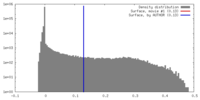
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yeast coatomer complex
| Entire | Name: Yeast coatomer complex |
|---|---|
| Components |
|
-Supramolecule #1000: Yeast coatomer complex
| Supramolecule | Name: Yeast coatomer complex / type: sample / ID: 1000 / Oligomeric state: one heteroheptamer / Number unique components: 7 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Cop1
| Macromolecule | Name: Cop1 / type: protein_or_peptide / ID: 1 / Name.synonym: alpha / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 136 KDa |
-Macromolecule #2: Sec26
| Macromolecule | Name: Sec26 / type: protein_or_peptide / ID: 2 / Name.synonym: beta / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 109 KDa |
-Macromolecule #3: Sec27
| Macromolecule | Name: Sec27 / type: protein_or_peptide / ID: 3 / Name.synonym: beta' / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 109 KDa |
-Macromolecule #4: Sec21
| Macromolecule | Name: Sec21 / type: protein_or_peptide / ID: 4 / Name.synonym: gamma / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 105 KDa |
-Macromolecule #5: Ret2
| Macromolecule | Name: Ret2 / type: protein_or_peptide / ID: 5 / Name.synonym: delta / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61 KDa |
-Macromolecule #6: Sec28
| Macromolecule | Name: Sec28 / type: protein_or_peptide / ID: 6 / Name.synonym: epsilon / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34 KDa |
-Macromolecule #7: Ret3
| Macromolecule | Name: Ret3 / type: protein_or_peptide / ID: 7 / Name.synonym: zeta / Number of copies: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22 KDa |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 0.005% NP40, 0.5 mM DTT |
| Staining | Type: NEGATIVE Details: Protein solution was adsorbed onto grids for 30 seconds before staining with 0.75% uranyl formate |
| Vitrification | Cryogen name: NITROGEN / Chamber temperature: 100 K / Instrument: OTHER / Method: Manual plunging into liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 100 K |
| Details | Low dose imaging, same specimen area imaged twice |
| Date | Mar 1, 2009 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 21 µm |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 51159 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: OTHER / Tilt angle max: 50 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 36.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Details: Final map is filtered to 36 Angstroms resolution |
|---|---|
| Final angle assignment | Details: SPIDER:theta 45 degrees, phi 45 degrees |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)