+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of holo-GmNifEN | ||||||||||||
 Map data Map data | CryoEM structure of holo-GmNifEN sharped with EMready | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | NITROGENASE COFACTOR MATURATION / PROTEIN BINDING / Metalloenzyme / Iron-Sulfur Cluster / METAL BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Geobacter metallireducens (bacteria) Geobacter metallireducens (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.86 Å | ||||||||||||
 Authors Authors | Paya Tormo L / Nguyen TQ / Fyfe C / Basbous H / Dobrzynska K / Echavarri-Erasun C / Martin L / Caserta G / Legrand P / Thorn A ...Paya Tormo L / Nguyen TQ / Fyfe C / Basbous H / Dobrzynska K / Echavarri-Erasun C / Martin L / Caserta G / Legrand P / Thorn A / Amara P / Schoehn G / Cherrier MV / Rubio LM / Nicolet Y | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2025 Journal: Nat Chem Biol / Year: 2025Title: Dynamics driving the precursor in NifEN scaffold during nitrogenase FeMo-cofactor assembly. Authors: Lucía Payá Tormo / Tu-Quynh Nguyen / Cameron Fyfe / Hind Basbous / Katarzyna Dobrzyńska / Carlos Echavarri-Erasun / Lydie Martin / Giorgio Caserta / Pierre Legrand / Andrea Thorn / ...Authors: Lucía Payá Tormo / Tu-Quynh Nguyen / Cameron Fyfe / Hind Basbous / Katarzyna Dobrzyńska / Carlos Echavarri-Erasun / Lydie Martin / Giorgio Caserta / Pierre Legrand / Andrea Thorn / Patricia Amara / Guy Schoehn / Mickaël V Cherrier / Luis M Rubio / Yvain Nicolet /    Abstract: Nitrogenase catalyzes atmospheric nitrogen fixation, a critical biological process that depends on an intricate organometallic cofactor assembled by a dedicated multiprotein system. Here we uncover ...Nitrogenase catalyzes atmospheric nitrogen fixation, a critical biological process that depends on an intricate organometallic cofactor assembled by a dedicated multiprotein system. Here we uncover the structural basis for the function of NifEN, the scaffold protein that mediates the final stages of cofactor biosynthesis before its incorporation into nitrogenase. High-resolution structural analyses reveal that the cofactor precursor initially binds at a surface docking site before being transferred into a specialized cavity for further maturation. This process involves dynamic structural rearrangements, including coordinated domain motions and partial unfolding, enabling the scaffold to alternate between open and closed states. Additionally, a rear channel extends to the precursor-binding cavity, likely facilitating the entry of the modifying components molybdenum and homocitrate. These findings illuminate the dynamic mechanisms underlying FeMo-cofactor assembly and underscore the functional divergence between NifEN, the biosynthetic scaffold, and NifDK, the catalytic component of nitrogenase. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52557.map.gz emd_52557.map.gz | 47.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52557-v30.xml emd-52557-v30.xml emd-52557.xml emd-52557.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_52557_fsc.xml emd_52557_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_52557.png emd_52557.png | 36.6 KB | ||
| Filedesc metadata |  emd-52557.cif.gz emd-52557.cif.gz | 7.6 KB | ||
| Others |  emd_52557_additional_1.map.gz emd_52557_additional_1.map.gz emd_52557_additional_2.map.gz emd_52557_additional_2.map.gz emd_52557_half_map_1.map.gz emd_52557_half_map_1.map.gz emd_52557_half_map_2.map.gz emd_52557_half_map_2.map.gz | 28.6 MB 53.5 MB 52.6 MB 52.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52557 http://ftp.pdbj.org/pub/emdb/structures/EMD-52557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52557 | HTTPS FTP |
-Validation report
| Summary document |  emd_52557_validation.pdf.gz emd_52557_validation.pdf.gz | 657.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_52557_full_validation.pdf.gz emd_52557_full_validation.pdf.gz | 657.3 KB | Display | |
| Data in XML |  emd_52557_validation.xml.gz emd_52557_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_52557_validation.cif.gz emd_52557_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52557 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52557 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52557 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52557 | HTTPS FTP |
-Related structure data
| Related structure data |  9i0gMC  9i0fC  9i0hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_52557.map.gz / Format: CCP4 / Size: 56.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52557.map.gz / Format: CCP4 / Size: 56.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of holo-GmNifEN sharped with EMready | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: CryoEM structure of holo-GmNifEN unsharped
| File | emd_52557_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of holo-GmNifEN unsharped | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: CryoEM structure of holo-GmNifEN sharped
| File | emd_52557_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of holo-GmNifEN sharped | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM structure of holo-GmNifEN
| File | emd_52557_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of holo-GmNifEN | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM structure of holo-GmNifEN
| File | emd_52557_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of holo-GmNifEN | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : nitrogenase iron-molybdenum cofactor biosynthesis protein NifEN
| Entire | Name: nitrogenase iron-molybdenum cofactor biosynthesis protein NifEN |
|---|---|
| Components |
|
-Supramolecule #1: nitrogenase iron-molybdenum cofactor biosynthesis protein NifEN
| Supramolecule | Name: nitrogenase iron-molybdenum cofactor biosynthesis protein NifEN type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (bacteria) Geobacter metallireducens (bacteria) |
| Molecular weight | Theoretical: 199.774 KDa |
-Macromolecule #1: Nitrogenase iron-molybdenum cofactor biosynthesis protein NifE
| Macromolecule | Name: Nitrogenase iron-molybdenum cofactor biosynthesis protein NifE type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (bacteria) / Strain: strain ATCC 53774 / DSM 7210 / GS-15 Geobacter metallireducens (bacteria) / Strain: strain ATCC 53774 / DSM 7210 / GS-15 |
| Molecular weight | Theoretical: 100.135594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKPDYYDVS ECETHEKGAP KFCKKSEPGE GAERSCAYDG ARVVLMPITD VIHLVHGPIA CAGNSWDNRG ARSSDSQLYR RGFTTEMLE NDVVFGGEKK LYRAILELAE RYEGQAKAMF VYATCVTAMT GDDVEAVCAA AGKKVAIPLI PVNTPGFIGD K NIGNRLAG ...String: MAKPDYYDVS ECETHEKGAP KFCKKSEPGE GAERSCAYDG ARVVLMPITD VIHLVHGPIA CAGNSWDNRG ARSSDSQLYR RGFTTEMLE NDVVFGGEKK LYRAILELAE RYEGQAKAMF VYATCVTAMT GDDVEAVCAA AGKKVAIPLI PVNTPGFIGD K NIGNRLAG EVLFKHVIGT AEPPVLGEYP INLIGEYNIA GDLWGMLPLF ERLGIQVLSC FSGDATFEEL RYAHRAKLNI II CSKSLTN LARKMQKNYG MPYLEESFYG MTDTAKALRD IARELDDAVG GLEKRIMQDR VEKLLEEEEA TCRERLAPYR ARL EGKRSV LFTGGVKTWS MVNALRELGV EILAAGTQNS TLEDFYRMKA LMHQDARIIE DTSSAGLLQV MYDKMPDLIV AGGK TKFLA LKTKTPFLDI NHGRSHPYAG YEGMVTFAKQ LDLTVNNPIW PVLNAKAPWE KTEEELTAAV ALAAGHARAC LDEDL KDST VKVPAKNATV NPQKNSPALG ATLAYLGIDQ MLALLHGAQG CSTFIRLQLS RHFKEPVALN STAMSEDTAI FGGWEN LKK GLKKVIEKFS PEVVGVMTSG LTETMGDDVR SAIVHFRQEY PEHDGVPVVW ASTPDYCGSL QEGYAATVEA IVRSVPE PG ETIPGQVTVL PGAHLTPADV EEVRELCEAF GLDPIIVPDI ANALDGHIDE TVSPLSTGGV SMARIRQAGQ SAATLFIG D SLAKAAEAMT ERCGMPSYGF TSLTGLAQVD RFMETLAAIA GRPIPEKFRR WRSRLMDAMV DSHYQFGLKK VTVALEGDN LKTLVNFLAG MGCEIQAAIA ATRVRGLDGL PARDIFVGDL EDLETAARGS DLIVANSNGR QAAAKLGIKA HLRAGLPVFD RLGAHQKMW VGYRGTMNLL FETANLFQAN AGEGQKLAHN UniProtKB: Nitrogenase iron-molybdenum cofactor biosynthesis protein NifE |
-Macromolecule #2: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 2 / Number of copies: 2 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #3: FeFe cofactor
| Macromolecule | Name: FeFe cofactor / type: ligand / ID: 3 / Number of copies: 2 / Formula: S5Q |
|---|---|
| Molecular weight | Theoretical: 747.356 Da |
| Chemical component information | 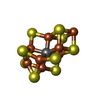 ChemComp-S5Q: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER/RHODIUM / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 30 mA | ||||||||
| Vitrification | Cryogen name: PROPANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-40 / Number grids imaged: 1 / Number real images: 2758 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 36000 |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Software | Name: PHENIX (ver. 1.21.1_5286:) |
| Details | AlphaFold Model of gmNifEN |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-9i0g: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















































