+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TRPML1 in complex with compound 4a | |||||||||
 Map data Map data | CryoSPARC sharpened masked filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of lysosome organization / calcium ion export / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / phagosome maturation / NAADP-sensitive calcium-release channel activity / iron ion transmembrane transporter activity / ligand-gated calcium channel activity / cellular response to pH / Transferrin endocytosis and recycling / iron ion transmembrane transport ...positive regulation of lysosome organization / calcium ion export / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / phagosome maturation / NAADP-sensitive calcium-release channel activity / iron ion transmembrane transporter activity / ligand-gated calcium channel activity / cellular response to pH / Transferrin endocytosis and recycling / iron ion transmembrane transport / monoatomic anion channel activity / TRP channels / sodium channel activity / monoatomic cation transport / autophagosome maturation / potassium channel activity / phagocytic cup / monoatomic cation channel activity / release of sequestered calcium ion into cytosol / cellular response to calcium ion / transferrin transport / cell projection / calcium ion transmembrane transport / calcium channel activity / phagocytic vesicle membrane / late endosome membrane / late endosome / protein homotetramerization / adaptive immune response / lysosome / receptor complex / endosome membrane / lysosomal membrane / intracellular membrane-bounded organelle / lipid binding / Golgi apparatus / nucleoplasm / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Reeks J / Mahajan P / Clark M / Cowan SR / Di Daniel E / Earl CP / Fisher S / Holvey RS / Jackson SM / Lloyd-Evans E ...Reeks J / Mahajan P / Clark M / Cowan SR / Di Daniel E / Earl CP / Fisher S / Holvey RS / Jackson SM / Lloyd-Evans E / Morgillo CM / Mortenson PN / O'Reilly M / Richardson CJ / Schopf P / Tams DM / Waller-Evans H / Ward SE / Whibley S / Williams PA / Johnson CN | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Enabling High Throughput Electron Cryo-microscopy for Structure-based Design Authors: Reeks J / Mahajan P / Clark M / Cowan SR / Di Daniel E / Earl CP / Fisher S / Holvey RS / Jackson SM / Lloyd-Evans E / Morgillo CM / Mortenson PN / O'Reilly M / Richardson CJ / Schopf P / ...Authors: Reeks J / Mahajan P / Clark M / Cowan SR / Di Daniel E / Earl CP / Fisher S / Holvey RS / Jackson SM / Lloyd-Evans E / Morgillo CM / Mortenson PN / O'Reilly M / Richardson CJ / Schopf P / Tams DM / Waller-Evans H / Ward SE / Whibley S / Williams PA / Johnson CN | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52245.map.gz emd_52245.map.gz | 54.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52245-v30.xml emd-52245-v30.xml emd-52245.xml emd-52245.xml | 30.3 KB 30.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_52245.png emd_52245.png | 126 KB | ||
| Masks |  emd_52245_msk_1.map emd_52245_msk_1.map | 783.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-52245.cif.gz emd-52245.cif.gz | 7.7 KB | ||
| Others |  emd_52245_additional_1.map.gz emd_52245_additional_1.map.gz emd_52245_additional_2.map.gz emd_52245_additional_2.map.gz emd_52245_half_map_1.map.gz emd_52245_half_map_1.map.gz emd_52245_half_map_2.map.gz emd_52245_half_map_2.map.gz | 68.9 MB 403 MB 725.7 MB 725.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52245 http://ftp.pdbj.org/pub/emdb/structures/EMD-52245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52245 | HTTPS FTP |
-Validation report
| Summary document |  emd_52245_validation.pdf.gz emd_52245_validation.pdf.gz | 704.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_52245_full_validation.pdf.gz emd_52245_full_validation.pdf.gz | 703.8 KB | Display | |
| Data in XML |  emd_52245_validation.xml.gz emd_52245_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_52245_validation.cif.gz emd_52245_validation.cif.gz | 25.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52245 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52245 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52245 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-52245 | HTTPS FTP |
-Related structure data
| Related structure data |  9hl6MC  9hj6C  9hj8C  9hl3C  9hl4C  9hl8C  9hlaC  9hlbC  9hlcC  9hldC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_52245.map.gz / Format: CCP4 / Size: 783.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52245.map.gz / Format: CCP4 / Size: 783.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC sharpened masked filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.6431 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_52245_msk_1.map emd_52245_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: CryoSPARC sharpened masked filtered map re-scaled to 0.5 A/pix
| File | emd_52245_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC sharpened masked filtered map re-scaled to 0.5 A/pix | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: CryoSPARC sharpened unmasked unfiltered map
| File | emd_52245_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC sharpened unmasked unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoSPARC non-uniform refinement half map 1
| File | emd_52245_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC non-uniform refinement half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoSPARC non-uniform refinement half map 2
| File | emd_52245_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC non-uniform refinement half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mucolipin-1
| Entire | Name: Mucolipin-1 |
|---|---|
| Components |
|
-Supramolecule #1: Mucolipin-1
| Supramolecule | Name: Mucolipin-1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 281 KDa |
-Macromolecule #1: Mucolipin-1
| Macromolecule | Name: Mucolipin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.518469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHHHG GSDYKDHDGD YKDHDIDYKD DDDKGGSGGS ENLYFQGPGT APAGPRGSET ERLLTPNPGY GTQAGPSPAP PTPPEEEDL RRRLKYFFMS PCDKFRAKGR KPCKLMLQVV KILVVTVQLI LFGLSNQLAV TFREENTIAF RHLFLLGYSD G ADDTFAAY ...String: MHHHHHHHHG GSDYKDHDGD YKDHDIDYKD DDDKGGSGGS ENLYFQGPGT APAGPRGSET ERLLTPNPGY GTQAGPSPAP PTPPEEEDL RRRLKYFFMS PCDKFRAKGR KPCKLMLQVV KILVVTVQLI LFGLSNQLAV TFREENTIAF RHLFLLGYSD G ADDTFAAY TREQLYQAIF HAVDQYLALP DVSLGRYAYV RGGGDPWTNG SGLALCQRYY HRGHVDPAND TFDIDPMVVT DC IQVDPPE RPPPPPSDDL TLLESSSSYK NLTLKFHKLV NVTIHFRLKT INLQSLINNE IPDCYTFSVL ITFDNKAHSG RIP ISLETQ AHIQECKHPS VFQHGDNSFR LLFDVVVILT CSLSFLLCAR SLLRGFLLQN EFVGFMWRQR GRVISLWERL EFVN GWYIL LVTSDVLTIS GTIMKIGIEA KNLASYDVCS ILLGTSTLLV WVGVIRYLTF FHNYNILIAT LRVALPSVMR FCCCV AVIY LGYCFCGWIV LGPYHVKFRS LSMVSECLFS LINGDDMFVT FAAMQAQQGR SSLVWLFSQL YLYSFISLFI YMVLSL FIA LITGAYDTIK HPGGAGAEES ELQAYIAQCQ DSPTSGKFRR GSGSACSLLC CCGRDPSEEH SLLVN UniProtKB: Mucolipin-1 |
-Macromolecule #3: PENTANE
| Macromolecule | Name: PENTANE / type: ligand / ID: 3 / Number of copies: 4 / Formula: LNK |
|---|---|
| Molecular weight | Theoretical: 72.149 Da |
| Chemical component information | 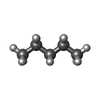 ChemComp-LNK: |
-Macromolecule #4: N-OCTANE
| Macromolecule | Name: N-OCTANE / type: ligand / ID: 4 / Number of copies: 8 / Formula: OCT |
|---|---|
| Molecular weight | Theoretical: 114.229 Da |
| Chemical component information |  ChemComp-OCT: |
-Macromolecule #5: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 5 / Number of copies: 4 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Macromolecule #6: HEXANE
| Macromolecule | Name: HEXANE / type: ligand / ID: 6 / Number of copies: 4 / Formula: HEX |
|---|---|
| Molecular weight | Theoretical: 86.175 Da |
| Chemical component information |  ChemComp-HEX: |
-Macromolecule #7: (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bi...
| Macromolecule | Name: (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate type: ligand / ID: 7 / Number of copies: 4 / Formula: EUJ |
|---|---|
| Molecular weight | Theoretical: 746.566 Da |
| Chemical component information | 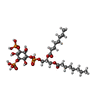 ChemComp-EUJ: |
-Macromolecule #8: 4-[[(3R)-3-[1-(4-chloranyl-2-fluoranyl-phenyl)piperidin-4-yl]-3-m...
| Macromolecule | Name: 4-[[(3R)-3-[1-(4-chloranyl-2-fluoranyl-phenyl)piperidin-4-yl]-3-methyl-2H-indol-1-yl]sulfonyl]-N,N-dimethyl-benzenesulfonamide type: ligand / ID: 8 / Number of copies: 4 / Formula: A1IV1 |
|---|---|
| Molecular weight | Theoretical: 592.145 Da |
-Macromolecule #9: water
| Macromolecule | Name: water / type: ligand / ID: 9 / Number of copies: 312 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number grids imaged: 1 / Average electron dose: 42.95 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: experimental model / Details: In house structure |
|---|---|
| Refinement | Space: REAL / Overall B value: 60 |
| Output model |  PDB-9hl6: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




























































