[English] 日本語
 Yorodumi
Yorodumi- EMDB-5217: Visualizing the structural changes of bacteriophage epsilon15 and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5217 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
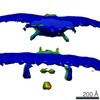
| Title | Visualizing the structural changes of bacteriophage epsilon15 and its Salmonella host during infection | |||||||||
 Map data Map data | Empty capsid attached to cell | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / infection / Salmonella / tomography / bacteriophage | |||||||||
| Biological species |   epsilon15 (virus) epsilon15 (virus) | |||||||||
| Method | subtomogram averaging / cryo EM | |||||||||
 Authors Authors | Chang JT / Schmid MF / Haase-Pettingell C / Weigele PR / King JA / Chiu W | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2010 Journal: J Mol Biol / Year: 2010Title: Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. Authors: Juan T Chang / Michael F Schmid / Cameron Haase-Pettingell / Peter R Weigele / Jonathan A King / Wah Chiu /  Abstract: The efficient mechanism by which double-stranded DNA bacteriophages deliver their chromosome across the outer membrane, cell wall, and inner membrane of Gram-negative bacteria remains obscure. ...The efficient mechanism by which double-stranded DNA bacteriophages deliver their chromosome across the outer membrane, cell wall, and inner membrane of Gram-negative bacteria remains obscure. Advances in single-particle electron cryomicroscopy have recently revealed details of the organization of the DNA injection apparatus within the mature virion for various bacteriophages, including epsilon15 (ɛ15) and P-SSP7. We have used electron cryotomography and three-dimensional subvolume averaging to capture snapshots of ɛ15 infecting its host Salmonella anatum. These structures suggest the following stages of infection. In the first stage, the tailspikes of ɛ15 attach to the surface of the host cell. Next, ɛ15's tail hub attaches to a putative cell receptor and establishes a tunnel through which the injection core proteins behind the portal exit the virion. A tube spanning the periplasmic space is formed for viral DNA passage, presumably from the rearrangement of core proteins or from cellular components. This tube would direct the DNA into the cytoplasm and protect it from periplasmic nucleases. Once the DNA has been injected into the cell, the tube and portal seals, and the empty bacteriophage remains at the cell surface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5217.map.gz emd_5217.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5217-v30.xml emd-5217-v30.xml emd-5217.xml emd-5217.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5217_1.png emd_5217_1.png | 102.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5217 http://ftp.pdbj.org/pub/emdb/structures/EMD-5217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5217 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5217.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5217.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Empty capsid attached to cell | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage epsilon15, Salmonella anatum
| Entire | Name: Bacteriophage epsilon15, Salmonella anatum |
|---|---|
| Components |
|
-Supramolecule #1000: Bacteriophage epsilon15, Salmonella anatum
| Supramolecule | Name: Bacteriophage epsilon15, Salmonella anatum / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: epsilon15
| Supramolecule | Name: epsilon15 / type: virus / ID: 1 / Name.synonym: epsilon15 / Sci species name: epsilon15 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: epsilon15 |
|---|---|
| Host (natural) | Organism:  Salmonella enterica subsp. enterica serovar Anatum (bacteria) Salmonella enterica subsp. enterica serovar Anatum (bacteria)synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Diameter: 700 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: 200 mesh copper grid |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 93 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Temperature | Min: 93 K / Max: 93 K / Average: 93 K |
| Specialist optics | Energy filter - Name: JEM3200FSC / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Sep 16, 2006 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Average electron dose: 65 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 9.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 20000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: JEOL 3200FSC CRYOHOLDER |
- Image processing
Image processing
| Final reconstruction | Details: The 3D reconstructions were performed with IMOD using gold as fiducial markers. Subvolumes, each containing a single particle, were computationally extracted from the reconstructions, ...Details: The 3D reconstructions were performed with IMOD using gold as fiducial markers. Subvolumes, each containing a single particle, were computationally extracted from the reconstructions, aligned to a reference model, and averaged together. |
|---|
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















