[English] 日本語
 Yorodumi
Yorodumi- EMDB-5214: Molecular architecture of the yeast TRAPPII tethering complex - form2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5214 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular architecture of the yeast TRAPPII tethering complex - form2 | |||||||||
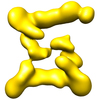
 Map data Map data | This is an image of a surface rendered top-view of TRAPPII form2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tethering complex / guanine nucleotide exchange factor | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 34.0 Å | |||||||||
 Authors Authors | Yip CK / Berscheminski J / Walz T | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2010 Journal: Nat Struct Mol Biol / Year: 2010Title: Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Authors: Calvin K Yip / Julia Berscheminski / Thomas Walz /  Abstract: Multisubunit tethering complexes participate in the process of vesicle tethering--the initial interaction between transport vesicles and their acceptor compartments. TRAPPII (named for transport ...Multisubunit tethering complexes participate in the process of vesicle tethering--the initial interaction between transport vesicles and their acceptor compartments. TRAPPII (named for transport protein particle II) is a highly conserved tethering complex that functions in the late Golgi apparatus and consists of all of the subunits of TRAPPI and three additional, specific subunits. We have purified native yeast TRAPPII and characterized its structure and subunit organization by single-particle EM. Our data show that the nine TRAPPII components form a core complex that dimerizes into a three-layered, diamond-shaped structure. The TRAPPI subunits assemble into TRAPPI complexes that form the outer layers. The three TRAPPII-specific subunits cap the ends of TRAPPI and form the middle layer, which is responsible for dimerization. TRAPPII binds the Ypt1 GTPase and probably uses the TRAPPI catalytic core to promote guanine nucleotide exchange. We discuss the implications of the structure of TRAPPII for coat interaction and TRAPPII-associated human pathologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5214.map.gz emd_5214.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5214-v30.xml emd-5214-v30.xml emd-5214.xml emd-5214.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5214_1.jpg emd_5214_1.jpg | 125 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5214 http://ftp.pdbj.org/pub/emdb/structures/EMD-5214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5214 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5214 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5214.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5214.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is an image of a surface rendered top-view of TRAPPII form2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Yeast TRAPPII complex
+Supramolecule #1000: Yeast TRAPPII complex
+Macromolecule #1: Bet3
+Macromolecule #2: Bet5
+Macromolecule #3: Trs20
+Macromolecule #4: Trs23
+Macromolecule #5: Trs31
+Macromolecule #6: Trs33
+Macromolecule #7: Kre11
+Macromolecule #8: Trs120
+Macromolecule #9: Trs130
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.005 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 10mM Tris-HCl 8.0, 150mM NaCl, 1mM Mg Acetate, 1mM imidazole, 20mM EGTA, 2mM beta-mercaptoethanol, 0.1% CHAPS |
| Staining | Type: NEGATIVE Details: Protein solution was adsorbed onto grids for 60 seconds before staining with 0.75% uranyl formate |
| Grid | Details: 400 mesh Quantifoil R1.2/1.3 overlaid with thin carbon |
| Vitrification | Cryogen name: NITROGEN / Chamber temperature: 100 K / Instrument: OTHER / Method: Manual plunging into liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 100 K |
| Details | Low dose imaging, same specimen area imaged twice |
| Date | Sep 4, 2009 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 21 µm / Number real images: 208 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 51159 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder.This holder operates in the temperature range from -175 C to ambient, and gives a resolution of at least 0.34 nm Specimen holder model: OTHER / Tilt angle max: 45 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected manually. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 34.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Details: Final map was filtered to 34 Angstroms / Number images used: 1686 |
| Final two d classification | Number classes: 2 |
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Software | Name:  Chimera Chimera | ||||||
| Details | PDBEntryID_givenInChain. Protocol: Rigid body. The full TRAPPI model was generated based on the two pdb entries listed and a third pdb file 3CUE. This model was manually docked into density map and further refined using Chimera. | ||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























