+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5196 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural map of MT 16-4 | |||||||||
 Map data Map data | Structural map of MT 16-4 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | microtubule / ultrastructure / Cryoelectron Microscopy Computer-Assisted Image Processing | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.5 Å | |||||||||
 Authors Authors | Sui H / Downing KH | |||||||||
 Citation Citation |  Journal: Structure / Year: 2010 Journal: Structure / Year: 2010Title: Structural basis of interprotofilament interaction and lateral deformation of microtubules. Authors: Haixin Sui / Kenneth H Downing /  Abstract: The diverse functions of microtubules require stiff structures possessing sufficient lateral flexibility to enable bending with high curvature. We used cryo-electron microscopy to investigate the ...The diverse functions of microtubules require stiff structures possessing sufficient lateral flexibility to enable bending with high curvature. We used cryo-electron microscopy to investigate the molecular basis for these critical mechanical properties. High-quality structural maps were used to build pseudoatomic models of microtubules containing 11-16 protofilaments, representing a wide range of lateral curvature. Protofilaments in all these microtubules were connected primarily via interprotofilament interactions between the M loops, and the H1'-S2 and H2-S3 loops. We postulate that the tolerance of the loop-loop interactions to lateral deformation provides the capacity for high-curvature bending without breaking. On the other hand, the local molecular architecture that surrounds these connecting loops contributes to the overall rigidity. Interprotofilament interactions in the seam region are similar to those in the normal helical regions, suggesting that the existence of the seam does not significantly affect the mechanical properties of microtubules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5196.map.gz emd_5196.map.gz | 17.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5196-v30.xml emd-5196-v30.xml emd-5196.xml emd-5196.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5196_1.jpg emd_5196_1.jpg | 162.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5196 http://ftp.pdbj.org/pub/emdb/structures/EMD-5196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5196 | HTTPS FTP |
-Validation report
| Summary document |  emd_5196_validation.pdf.gz emd_5196_validation.pdf.gz | 77.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5196_full_validation.pdf.gz emd_5196_full_validation.pdf.gz | 77 KB | Display | |
| Data in XML |  emd_5196_validation.xml.gz emd_5196_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5196 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5196 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5196 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5196 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5196.map.gz / Format: CCP4 / Size: 17.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5196.map.gz / Format: CCP4 / Size: 17.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structural map of MT 16-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
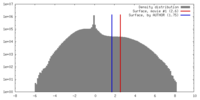
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The microtubule containing 16 protofilaments
| Entire | Name: The microtubule containing 16 protofilaments |
|---|---|
| Components |
|
-Supramolecule #1000: The microtubule containing 16 protofilaments
| Supramolecule | Name: The microtubule containing 16 protofilaments / type: sample / ID: 1000 Oligomeric state: The 16-protofilament microtubule forms a helical structure Number unique components: 2 |
|---|
-Macromolecule #1: microtubule with 16 protofilaments
| Macromolecule | Name: microtubule with 16 protofilaments / type: protein_or_peptide / ID: 1 / Name.synonym: MT 16-4 Details: Microtubule with 16 protofilaments and 4-start helical structure Oligomeric state: Dimer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 50 MDa / Theoretical: 50 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 / Details: 25mM Pipes, 25mM NaCl, 2mM MgCl2, 1mM EGTA |
|---|---|
| Grid | Details: 300 mesh copper grid covered with home-made carbon films |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 93 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Home made plunger / Method: Blot for 2 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 4000EX |
|---|---|
| Temperature | Average: 105 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 400,000 times magnification Legacy - Electron beam tilt params: 0 |
| Date | Jul 12, 2007 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 6.35 µm / Number real images: 425 / Average electron dose: 15 e/Å2 Details: The micrographs were digitized with a customized robotic scanning system that uses a Nikon Coolpix 8000 scanner Bits/pixel: 16 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 400 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | With addition of Taxol or Taxotere for microtubule stabilization |
|---|---|
| CTF correction | Details: Each micrograph |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 10.5 Å / Resolution method: OTHER / Software - Name: SPIDER |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid Body. A homology model was generated for the missing region in 1JFF from residues 35 to 60 in alpha-tubulin (by N. Banavali). The tubulin dimer structure was fitted into the density maps using the rigid body fitting function in Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)