+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5176 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
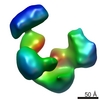
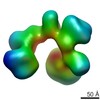
| Title | Frozen hydrated map of the yeast TFIID-TFIIA-Rap1-DNA complex | |||||||||
 Map data Map data | Yeast TFIID in complex with Rap1 and DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription / general transcription factor / TFIID / transcription initiation / transciption activator / TFIIA / Rap1 | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 24.5 Å | |||||||||
 Authors Authors | Papai G / Tripathi MK / Ruhlmann C / Layer JH / Weil PA / Schultz P | |||||||||
 Citation Citation |  Journal: Nature / Year: 2010 Journal: Nature / Year: 2010Title: TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Authors: Gabor Papai / Manish K Tripathi / Christine Ruhlmann / Justin H Layer / P Anthony Weil / Patrick Schultz /  Abstract: Transcription of eukaryotic messenger RNA (mRNA) encoding genes by RNA polymerase II (Pol II) is triggered by the binding of transactivating proteins to enhancer DNA, which stimulates the recruitment ...Transcription of eukaryotic messenger RNA (mRNA) encoding genes by RNA polymerase II (Pol II) is triggered by the binding of transactivating proteins to enhancer DNA, which stimulates the recruitment of general transcription factors (TFIIA, B, D, E, F, H) and Pol II on the cis-linked promoter, leading to pre-initiation complex formation and transcription. In TFIID-dependent activation pathways, this general transcription factor containing TATA-box-binding protein is first recruited on the promoter through interaction with activators and cooperates with TFIIA to form a committed pre-initiation complex. However, neither the mechanisms by which activation signals are communicated between these factors nor the structural organization of the activated pre-initiation complex are known. Here we used cryo-electron microscopy to determine the architecture of nucleoprotein complexes composed of TFIID, TFIIA, the transcriptional activator Rap1 and yeast enhancer-promoter DNA. These structures revealed the mode of binding of Rap1 and TFIIA to TFIID, as well as a reorganization of TFIIA induced by its interaction with Rap1. We propose that this change in position increases the exposure of TATA-box-binding protein within TFIID, consequently enhancing its ability to interact with the promoter. A large Rap1-dependent DNA loop forms between the activator-binding site and the proximal promoter region. This loop is topologically locked by a TFIIA-Rap1 protein bridge that folds over the DNA. These results highlight the role of TFIIA in transcriptional activation, define a molecular mechanism for enhancer-promoter communication and provide structural insights into the pathways of intramolecular communication that convey transcription activation signals through the TFIID complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5176.map.gz emd_5176.map.gz | 10.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5176-v30.xml emd-5176-v30.xml emd-5176.xml emd-5176.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5176.png emd_5176.png | 174.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5176 http://ftp.pdbj.org/pub/emdb/structures/EMD-5176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5176 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5176.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5176.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast TFIID in complex with Rap1 and DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HA-tag purified yeast TFIID
| Entire | Name: HA-tag purified yeast TFIID |
|---|---|
| Components |
|
-Supramolecule #1000: HA-tag purified yeast TFIID
| Supramolecule | Name: HA-tag purified yeast TFIID / type: sample / ID: 1000 / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1 MDa |
-Macromolecule #1: Rap1
| Macromolecule | Name: Rap1 / type: protein_or_peptide / ID: 1 / Name.synonym: Rap1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #2: TFIID
| Macromolecule | Name: TFIID / type: protein_or_peptide / ID: 2 / Name.synonym: TFIID / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.165 mg/mL |
|---|---|
| Buffer | pH: 7.9 / Details: 10 mM Tris-HCl pH 7.9, 300 mM KOAc and 5 mM MgCl2 |
| Grid | Details: 300 mesh Cu/Rh holey grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 80 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Average: 81 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 5.1 µm / Number real images: 90 / Average electron dose: 16 e/Å2 / Od range: 1.4 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38950 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.5 Å / Resolution method: OTHER / Software - Name: Imagic, Spider Details: The map was reconstructed from 3-D MSA separated dataset. Number images used: 26901 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















