[English] 日本語
 Yorodumi
Yorodumi- EMDB-51700: CryoEM structure of Gs-coupled GPBAR with small molecule agonist P395 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Gs-coupled GPBAR with small molecule agonist P395 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G-protein coupled receptor / transmembrane protein / G protein signaling / bile acid-binding. / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled bile acid receptor activity / adenylate cyclase-activating G protein-coupled bile acid receptor signaling pathway / positive regulation of cholangiocyte proliferation / cellular response to bile acid / Class A/1 (Rhodopsin-like receptors) / regulation of bicellular tight junction assembly / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding ...G protein-coupled bile acid receptor activity / adenylate cyclase-activating G protein-coupled bile acid receptor signaling pathway / positive regulation of cholangiocyte proliferation / cellular response to bile acid / Class A/1 (Rhodopsin-like receptors) / regulation of bicellular tight junction assembly / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / electron transport chain / bone development / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / cell population proliferation / receptor complex / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / Resolution: 2.5 Å | |||||||||
 Authors Authors | Frechard A / Brooks I | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2025 Journal: Sci Rep / Year: 2025Title: Deciphering molecular determinants of GPBAR1-Gs protein interactions by HDX-MS and cryo-EM. Authors: Jérôme Castel / Thomas Botzanowski / Ieva Brooks / Alexandre Frechard / Gilbert Bey / Marine Schroeter / Elise Del Nero / François Debaene / Fabrice Ciesielski / Denis Zeyer / Sarah ...Authors: Jérôme Castel / Thomas Botzanowski / Ieva Brooks / Alexandre Frechard / Gilbert Bey / Marine Schroeter / Elise Del Nero / François Debaene / Fabrice Ciesielski / Denis Zeyer / Sarah Cianferani / Renaud Morales /  Abstract: Many physiological processes are dependent on G protein-coupled receptors (GPCRs), the biggest family of human membrane proteins and a significant class of therapeutic targets. Once activated by ...Many physiological processes are dependent on G protein-coupled receptors (GPCRs), the biggest family of human membrane proteins and a significant class of therapeutic targets. Once activated by external stimuli, GPCRs use G proteins and arrestins as transducers to generate second messengers and trigger downstream signaling, leading to diverse signaling profiles. The G protein-coupled bile acid receptor 1 (GPBAR1, also known as Takeda G protein-coupled receptor 5, TGR5) is a class A bile acid membrane receptor that regulates energy homeostasis and glucose and lipid metabolism. GPBAR1/Gs protein interactions are implicated in the prevention of diabetes and the reduction of inflammatory responses, making GPBAR1 a potential therapeutic target for metabolic disorders. Here, we present combined hydrogen/deuterium exchange mass spectrometry (HDX-MS) and cryo-electron microscopy (cryo-EM) to identify the molecular determinants of GPBAR1 conformational dynamics upon G protein binding. Thanks to extensive optimization, we achieved over 75% sequence coverage by HDX-MS of a complete GPCR complex and a 2.5 Å resolution structure by cryo-EM, both of which are state-of-the-art. Altogether, our results provide information on the under-investigated GPBAR1 binding mode to its cognate G protein, pinpointing the synergic and powerful combination of higher cryo-EM and lower HDX-MS resolution techniques to dissect GPCR/G protein binding characteristics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51700.map.gz emd_51700.map.gz | 96.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51700-v30.xml emd-51700-v30.xml emd-51700.xml emd-51700.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51700_fsc.xml emd_51700_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_51700.png emd_51700.png | 52 KB | ||
| Filedesc metadata |  emd-51700.cif.gz emd-51700.cif.gz | 7.2 KB | ||
| Others |  emd_51700_half_map_1.map.gz emd_51700_half_map_1.map.gz emd_51700_half_map_2.map.gz emd_51700_half_map_2.map.gz | 81.2 MB 81.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51700 http://ftp.pdbj.org/pub/emdb/structures/EMD-51700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51700 | HTTPS FTP |
-Related structure data
| Related structure data |  9gyoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.731 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51700_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
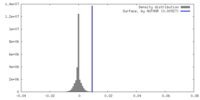
| Density Histograms |
-Half map: #1
| File | emd_51700_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
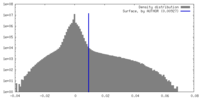
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the TGR5 with Gs protein heterotrimer, nanobody 35 and...
| Entire | Name: Complex of the TGR5 with Gs protein heterotrimer, nanobody 35 and small molecule agonist P395 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the TGR5 with Gs protein heterotrimer, nanobody 35 and...
| Supramolecule | Name: Complex of the TGR5 with Gs protein heterotrimer, nanobody 35 and small molecule agonist P395 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit...
| Macromolecule | Name: Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.385246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG AGESGKSTIV KQMRILHVNG FNGDSEKATK VQDIKNNLKE AIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR ACYERSNEYQ LIDCAQYFLD K IDVIKQAD ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RATHRLLLLG AGESGKSTIV KQMRILHVNG FNGDSEKATK VQDIKNNLKE AIETIVAAM SNLVPPVELA NPENQFRVDY ILSVMNVPDF DFPPEFYEHA KALWEDEGVR ACYERSNEYQ LIDCAQYFLD K IDVIKQAD YVPSDQDLLR CRVLTSGIFE TKFQVDKVNF HMFDVGGQRD ERRKWIQCFN DVTAIIFVVA SSSYNMVIRE DN QTNRLQE ALNLFKSIWN NRWLRTISVI LFLNKQDLLA EKVLAGKSKI EDYFPEFARY TTPEDATPEP GEDPRVTRAK YFI RDEFLR ISTASGDGRH YCYPHFTCAV DTENIRRVFN DCRDIIQRMH LRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.983127 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKYLLPTAA AGLLLLAAQP AMAQVQLQES GGGLVQPGGS LRLSCAASGF TFSNYKMNWV RQAPGKGLEW VSDISQSGAS ISYTGSVKG RFTISRDNAK NTLYLQMNSL KPEDTAVYYC ARCPAPFTRD CFDVTSTTYA YRGQGTQVTV SSHHHHHH |
-Macromolecule #5: Soluble cytochrome b562,G-protein coupled bile acid receptor 1
| Macromolecule | Name: Soluble cytochrome b562,G-protein coupled bile acid receptor 1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.883793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKLEVLFQ GPGSADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMTPNSTGEV PSPIPKGALG L SLALASLI ...String: MKTIIALSYI FCLVFADYKD DDDKLEVLFQ GPGSADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMTPNSTGEV PSPIPKGALG L SLALASLI ITANLLLALG IAWDRRLRSP PAGCFFLSLL LAGLLTGLAL PTLPGLWNQS RRGYWSCLLV YLAPNFSFLS LL ANLLLVH GERYMAVLRP LQPPGSIRLA LLLTWAGPLL FASLPALGWN HWTPGANCSS QAIFPAPYLY LEVYGLLLPA VGA AAFLSV RVLATAHRQL QDICRLERAV CRDEPSALAR ALTWRQARAQ AGAMLLFGLC WGPYVATLLL SVLAYEQRPP LGPG TLLSL LSLGSASAAA VPVAMGLGDQ RYTAPWRAAA QRCLQGLWGR ASRDSPGPSI AYHPSSQSSV DLDLN UniProtKB: Soluble cytochrome b562, G-protein coupled bile acid receptor 1 |
-Macromolecule #6: 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5...
| Macromolecule | Name: 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide type: ligand / ID: 6 / Number of copies: 1 / Formula: FWX |
|---|---|
| Molecular weight | Theoretical: 395.498 Da |
| Chemical component information | 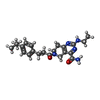 ChemComp-FWX: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 3 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)