[English] 日本語
 Yorodumi
Yorodumi- EMDB-51339: E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DN... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | DNA gyrase / type II topoisomerase / moxifloxacin / DNA crossover / ISOMERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of DNA-templated DNA replication / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex / DNA negative supercoiling activity / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / ATP-dependent activity, acting on DNA / DNA-templated DNA replication / chromosome / response to xenobiotic stimulus ...negative regulation of DNA-templated DNA replication / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex / DNA negative supercoiling activity / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / ATP-dependent activity, acting on DNA / DNA-templated DNA replication / chromosome / response to xenobiotic stimulus / response to antibiotic / DNA-templated transcription / DNA binding / ATP binding / metal ion binding / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   Escherichia phage Mu (virus) Escherichia phage Mu (virus) | ||||||||||||
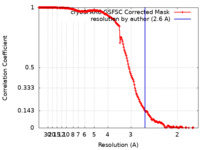
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | ||||||||||||
 Authors Authors | Ghilarov D / Heddle JG / Pabis M | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Poland, 3 items Poland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structural basis of chiral wrap and T-segment capture by DNA gyrase. Authors: Elizabeth Michalczyk / Zuzanna Pakosz-Stępień / Jonathon D Liston / Olivia Gittins / Marta Pabis / Jonathan G Heddle / Dmitry Ghilarov /   Abstract: Type II topoisomerase DNA gyrase transduces the energy of ATP hydrolysis into the negative supercoiling of DNA. The postulated catalytic mechanism involves stabilization of a chiral DNA loop followed ...Type II topoisomerase DNA gyrase transduces the energy of ATP hydrolysis into the negative supercoiling of DNA. The postulated catalytic mechanism involves stabilization of a chiral DNA loop followed by the passage of the T-segment through the temporarily cleaved G-segment resulting in sign inversion. The molecular basis for this is poorly understood as the chiral loop has never been directly observed. We have obtained high-resolution cryoEM structures of gyrase with chirally wrapped 217 bp DNA with and without the fluoroquinolone moxifloxacin (MFX). Each structure constrains a positively supercoiled figure-of-eight DNA loop stabilized by a GyrA β-pinwheel domain which has the structure of a flat disc. By comparing the catalytic site of the native drug-free and MFX-bound gyrase structures both of which contain a single metal ion, we demonstrate that the enzyme is observed in a native precatalytic state. Our data imply that T-segment trapping is not dependent on the dimerization of the ATPase domains which appears to only be possible after strand passage has taken place. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51339.map.gz emd_51339.map.gz | 256.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51339-v30.xml emd-51339-v30.xml emd-51339.xml emd-51339.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51339_fsc.xml emd_51339_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_51339.png emd_51339.png | 113.7 KB | ||
| Filedesc metadata |  emd-51339.cif.gz emd-51339.cif.gz | 8.2 KB | ||
| Others |  emd_51339_half_map_1.map.gz emd_51339_half_map_1.map.gz emd_51339_half_map_2.map.gz emd_51339_half_map_2.map.gz | 475.2 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51339 http://ftp.pdbj.org/pub/emdb/structures/EMD-51339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51339 | HTTPS FTP |
-Related structure data
| Related structure data |  9ggqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51339.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51339.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
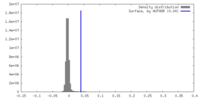
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51339_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
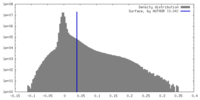
| Density Histograms |
-Half map: #1
| File | emd_51339_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
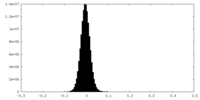
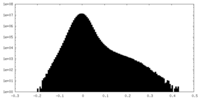
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Escherichia coli DNA gyrase with 217 bp linear...
| Entire | Name: Ternary complex of Escherichia coli DNA gyrase with 217 bp linear DNA fragment and moxifloxacin |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Escherichia coli DNA gyrase with 217 bp linear...
| Supramolecule | Name: Ternary complex of Escherichia coli DNA gyrase with 217 bp linear DNA fragment and moxifloxacin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA gyrase subunit A
| Macromolecule | Name: DNA gyrase subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA topoisomerase (ATP-hydrolysing) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.854305 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SSDLAREITP VNIEEELKSS YLDYAMSVIV GRALPDVRDG LKPVHRRVLY AMNVLGNDWN KAYKKSARVV GDVIGKYHPH GDSAVYDTI VRMAQPFSLR YMLVDGQGNF GSIDGDSAAA MR(PTR)TEIRLAK IAHELMADLE KETVDFVDNY DGTEKIP DV MPTKIPNLLV ...String: SSDLAREITP VNIEEELKSS YLDYAMSVIV GRALPDVRDG LKPVHRRVLY AMNVLGNDWN KAYKKSARVV GDVIGKYHPH GDSAVYDTI VRMAQPFSLR YMLVDGQGNF GSIDGDSAAA MR(PTR)TEIRLAK IAHELMADLE KETVDFVDNY DGTEKIP DV MPTKIPNLLV NGSSGIAVGM ATNIPPHNLT EVINGCLAYI DDEDISIEGL MEHIPGPDFP TAAIINGRRG IEEAYRTG R GKVYIRARAE VEVDAKTGRE TIIVHEIPYQ VNKARLIEKI AELVKEKRVE GISALRDESD KDGMRIVIEV KRDAVGEVV LNNLYSQTQL QVSFGINMVA LHHGQPKIMN LKDIIAAFVR HRREVVTRRT IFELRKARDR AHILEALAVA LANIDPIIEL IRHAPTPAE AKTALVANPW QLGNVAAMLE RAGDDAARPE WLEPEFGVRD GLYYLTEQQA QAILDLRLQK LTGLEHEKLL D EYKELLDQ IAELLRILGS ADRLMEVIRE ELELVREQFG DKRRTEITAN SADINLEDLI TQEDVVVTLS HQGYVKYQPL SE YEAQRRG GKGKSAARIK EEDFIDRLLV ANTHDHILCF SSRGRVYSMK VYQLPEATRG ARGRPIVNLL PLEQDERITA ILP VTEFEE GVKVFMATAN GTVKKTVLTE FNRLRTAGKV AIKLVDGDEL IGVDLTSGED EVMLFSAEGK VVRFKESSVR AMGC NTTGV RGIRLGEGDK VVSLIVPRGD GAILTATQNG YGKRTAVAEY PTKSRATKGV ISIKVTERNG LVVGAVQVDD CDQIM MITD AGTLVRTRVS EISIVGRNTQ GVILIRTAED ENVVGLQRVA EPVDEEDLDT IDGSAAEGDD EIAPEVDVDD EPEEEL EVL FQ UniProtKB: DNA gyrase subunit A |
-Macromolecule #2: DNA gyrase subunit B
| Macromolecule | Name: DNA gyrase subunit B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA topoisomerase (ATP-hydrolysing) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90.891734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSNSYDSSS IKVLKGLDAV RKRPGMYIGD TDDGTGLHHM VFEVVDNAID EALAGHCKEI IVTIHADNSV SVQDDGRGIP TGIHPEEGV SAAEVIMTVL HAGGKFDDNS YKVSGGLHGV GVSVVNALSQ KLELVIQREG KIHRQIYEHG VPQAPLAVTG E TEKTGTMV ...String: GPSNSYDSSS IKVLKGLDAV RKRPGMYIGD TDDGTGLHHM VFEVVDNAID EALAGHCKEI IVTIHADNSV SVQDDGRGIP TGIHPEEGV SAAEVIMTVL HAGGKFDDNS YKVSGGLHGV GVSVVNALSQ KLELVIQREG KIHRQIYEHG VPQAPLAVTG E TEKTGTMV RFWPSLETFT NVTEFEYEIL AKRLRELSFL NSGVSIRLRD KRDGKEDHFH YEGGIKAFVE YLNKNKTPIH PN IFYFSTE KDGIGVEVAL QWNDGFQENI YCFTNNIPQR DGGTHLAGFR AAMTRTLNAY MDKEGYSKKA KVSATGDDAR EGL IAVVSV KVPDPKFSSQ TKDKLVSSEV KSAVEQQMNE LLAEYLLENP TDAKIVVGKI IDAARAREAA RRAREMTRRK GALD LAGLP GKLADCQERD PALSELYLVE GDSAGGSAKQ GRNRKNQAIL PLKGKILNVE KARFDKMLSS QEVATLITAL GCGIG RDEY NPDKLRYHSI IIMTDADVDG SHIRTLLLTF FYRQMPEIVE RGHVYIAQPP LYKVKKGKQE QYIKDDEAMD QYQISI ALD GATLHTNASA PALAGEALEK LVSEYNATQK MINRMERRYP KAMLKELIYQ PTLTEADLSD EQTVTRWVNA LVSELND KE QHGSQWKFDV HTNAEQNLFE PIVRVRTHGV DTDYPLDHEF ITGGEYRRIC TLGEKLRGLL EEDAFIERGE RRQPVASF E QALDWLVKES RRGLSIQRYK GLGEMNPEQL WETTMDPESR RMLRVTVKDA IAADQLFTTL MGDAVEPRRA FIEENALKA ANIDIENLYF Q UniProtKB: DNA gyrase subunit B |
-Macromolecule #3: Mu217R
| Macromolecule | Name: Mu217R / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Escherichia phage Mu (virus) Escherichia phage Mu (virus) |
| Molecular weight | Theoretical: 65.926109 KDa |
| Sequence | String: (DT)(DT)(DC)(DC)(DT)(DG)(DC)(DG)(DC)(DG) (DT)(DC)(DC)(DT)(DT)(DA)(DT)(DA)(DT)(DG) (DG)(DG)(DG)(DC)(DT)(DG)(DG)(DA)(DT) (DA)(DA)(DG)(DA)(DC)(DC)(DG)(DG)(DG)(DG) (DA) (DA)(DC)(DA)(DG)(DG)(DA) ...String: (DT)(DT)(DC)(DC)(DT)(DG)(DC)(DG)(DC)(DG) (DT)(DC)(DC)(DT)(DT)(DA)(DT)(DA)(DT)(DG) (DG)(DG)(DG)(DC)(DT)(DG)(DG)(DA)(DT) (DA)(DA)(DG)(DA)(DC)(DC)(DG)(DG)(DG)(DG) (DA) (DA)(DC)(DA)(DG)(DG)(DA)(DA)(DA) (DT)(DT)(DT)(DT)(DT)(DG)(DA)(DT)(DG)(DC) (DC)(DA) (DT)(DC)(DA)(DG)(DA)(DA)(DA) (DA)(DC)(DG)(DC)(DG)(DT)(DC)(DA)(DG)(DC) (DG)(DC)(DC) (DG)(DC)(DT)(DC)(DT)(DG) (DA)(DG)(DG)(DC)(DA)(DA)(DT)(DA)(DA)(DA) (DC)(DA)(DG)(DA) (DA)(DT)(DC)(DA)(DG) (DG)(DC)(DA)(DT)(DA)(DA)(DA)(DA)(DT)(DC) (DA)(DC)(DC)(DC)(DG) (DC)(DA)(DC)(DA) (DG)(DA)(DT)(DT)(DT)(DT)(DT)(DT)(DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG) (DC)(DC)(DA) (DC)(DG)(DG)(DG)(DA)(DT)(DT)(DT)(DT)(DT) (DA)(DA)(DA)(DC)(DC)(DG)(DG) (DT)(DA) (DT)(DT)(DT)(DA)(DA)(DC)(DG)(DG)(DT)(DG) (DT)(DA)(DT)(DG)(DA)(DA)(DT)(DC) (DC) (DC)(DG)(DT)(DT)(DT)(DT)(DA)(DT)(DC)(DT) (DT)(DC)(DC)(DT)(DT)(DT)(DC)(DA)(DC) (DT)(DT)(DT)(DC)(DT)(DT)(DT)(DC)(DT)(DC) (DC)(DA)(DG)(DT) GENBANK: GENBANK: M74911.1 |
-Macromolecule #4: Mu217F
| Macromolecule | Name: Mu217F / type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Escherichia phage Mu (virus) Escherichia phage Mu (virus) |
| Molecular weight | Theoretical: 66.202258 KDa |
| Sequence | String: (DA)(DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DA) (DG)(DA)(DA)(DA)(DG)(DT)(DG)(DA)(DA)(DA) (DG)(DG)(DA)(DA)(DG)(DA)(DT)(DA)(DA) (DA)(DA)(DC)(DG)(DG)(DG)(DA)(DT)(DT)(DC) (DA) (DT)(DA)(DC)(DA)(DC)(DC) ...String: (DA)(DC)(DT)(DG)(DG)(DA)(DG)(DA)(DA)(DA) (DG)(DA)(DA)(DA)(DG)(DT)(DG)(DA)(DA)(DA) (DG)(DG)(DA)(DA)(DG)(DA)(DT)(DA)(DA) (DA)(DA)(DC)(DG)(DG)(DG)(DA)(DT)(DT)(DC) (DA) (DT)(DA)(DC)(DA)(DC)(DC)(DG)(DT) (DT)(DA)(DA)(DA)(DT)(DA)(DC)(DC)(DG)(DG) (DT)(DT) (DT)(DA)(DA)(DA)(DA)(DA)(DT) (DC)(DC)(DC)(DG)(DT)(DG)(DG)(DC)(DG)(DC) (DG)(DT)(DT) (DT)(DT)(DA)(DA)(DA)(DA) (DA)(DA)(DT)(DC)(DT)(DG)(DT)(DG)(DC)(DG) (DG)(DG)(DT)(DG) (DA)(DT)(DT)(DT)(DT) (DA)(DT)(DG)(DC)(DC)(DT)(DG)(DA)(DT)(DT) (DC)(DT)(DG)(DT)(DT) (DT)(DA)(DT)(DT) (DG)(DC)(DC)(DT)(DC)(DA)(DG)(DA)(DG)(DC) (DG)(DG)(DC)(DG)(DC)(DT) (DG)(DA)(DC) (DG)(DC)(DG)(DT)(DT)(DT)(DT)(DC)(DT)(DG) (DA)(DT)(DG)(DG)(DC)(DA)(DT) (DC)(DA) (DA)(DA)(DA)(DA)(DT)(DT)(DT)(DC)(DC)(DT) (DG)(DT)(DT)(DC)(DC)(DC)(DC)(DG) (DG) (DT)(DC)(DT)(DT)(DA)(DT)(DC)(DC)(DA)(DG) (DC)(DC)(DC)(DC)(DA)(DT)(DA)(DT)(DA) (DA)(DG)(DG)(DA)(DC)(DG)(DC)(DG)(DC)(DA) (DG)(DG)(DA)(DA) GENBANK: GENBANK: M74911.1 |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrol...
| Macromolecule | Name: 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid type: ligand / ID: 6 / Number of copies: 2 / Formula: MFX |
|---|---|
| Molecular weight | Theoretical: 401.431 Da |
| Chemical component information |  ChemComp-MFX: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 8 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)