+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | yeast TFIIIC TauA subcomplex bound to a tRNA gene | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | A-box / tRNA gene / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA Polymerase III Transcription Initiation From Type 2 Promoter / transcription initiation at RNA polymerase III promoter / phosphatase activity / transcription by RNA polymerase III / protein localization to chromatin / mitochondrion / DNA binding / nucleoplasm ...5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA Polymerase III Transcription Initiation From Type 2 Promoter / transcription initiation at RNA polymerase III promoter / phosphatase activity / transcription by RNA polymerase III / protein localization to chromatin / mitochondrion / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Seifert-Davila W / Girbig M / Hauptmann L / Hoffmann T / Eustermann S / Mueller C / Chaban A / Duss O / Baudin F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2025 Journal: Nucleic Acids Res / Year: 2025Title: Structural and kinetic insights into tRNA promoter engagement by yeast general transcription factor TFIIIC. Authors: Wolfram Seifert-Dávila / Anastasiia Chaban / Florence Baudin / Mathias Girbig / Luis Hauptmann / Thomas Hoffmann / Olivier Duss / Sebastian Eustermann / Christoph W Müller /  Abstract: Transcription of transfer RNA (tRNA) genes by RNA polymerase (Pol) III requires the general transcription factor IIIC (TFIIIC), which recognizes intragenic A-box and B-box DNA motifs of type II ...Transcription of transfer RNA (tRNA) genes by RNA polymerase (Pol) III requires the general transcription factor IIIC (TFIIIC), which recognizes intragenic A-box and B-box DNA motifs of type II gene promoters. However, the underlying mechanism has remained elusive, in part due to missing structural information for A-box recognition. In this study, we use single-particle cryogenic electron microscopy (cryo-EM) and single-molecule fluorescence resonance energy transfer (smFRET) to reveal structural and real-time kinetic insights into how the 520-kDa yeast TFIIIC complex engages A-box and B-box DNA motifs in the context of a tRNA gene promoter. Cryo-EM structures of τA and τB subcomplexes bound to the A-box and B-box were obtained at 3.7 and 2.5 Å resolution, respectively, while cryo-EM single-particle mapping determined the specific distance and relative orientation of the τA and τB subcomplexes revealing a fully engaged state of TFIIIC. smFRET experiments show that overall recruitment and residence times of TFIIIC on a tRNA gene are primarily governed by B-box recognition, while footprinting experiments suggest a key role of τA and the A-box in TFIIIB and Pol III recruitment following TFIIIC recognition of type II promoters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51231.map.gz emd_51231.map.gz | 98.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51231-v30.xml emd-51231-v30.xml emd-51231.xml emd-51231.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_51231.png emd_51231.png | 102 KB | ||
| Filedesc metadata |  emd-51231.cif.gz emd-51231.cif.gz | 8.4 KB | ||
| Others |  emd_51231_half_map_1.map.gz emd_51231_half_map_1.map.gz emd_51231_half_map_2.map.gz emd_51231_half_map_2.map.gz | 98.6 MB 98.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51231 http://ftp.pdbj.org/pub/emdb/structures/EMD-51231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51231 | HTTPS FTP |
-Related structure data
| Related structure data |  9gckMC  9gc3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51231.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51231.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
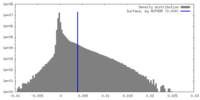
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_51231_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51231_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast TFIIIC TauA monomer bound to a tRNA gene
| Entire | Name: Yeast TFIIIC TauA monomer bound to a tRNA gene |
|---|---|
| Components |
|
-Supramolecule #1: Yeast TFIIIC TauA monomer bound to a tRNA gene
| Supramolecule | Name: Yeast TFIIIC TauA monomer bound to a tRNA gene / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Transcription factor tau
| Supramolecule | Name: Transcription factor tau / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transcription factor tau 138 kDa subunit
| Macromolecule | Name: Transcription factor tau 138 kDa subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 136.6025 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVLTIYPDEL VQIVSDKIAS NKGKITLNQL WDISGKYFDL SDKKVKQFVL SCVILKKDIE VYCDGAITTK NVTDIIGDAN HSYSVGITE DSLWTLLTGY TKKESTIGNS AFELLLEVAK SGEKGINTMD LAQVTGQDPR SVTGRIKKIN HLLTSSQLIY K GHVVKQLK ...String: MVLTIYPDEL VQIVSDKIAS NKGKITLNQL WDISGKYFDL SDKKVKQFVL SCVILKKDIE VYCDGAITTK NVTDIIGDAN HSYSVGITE DSLWTLLTGY TKKESTIGNS AFELLLEVAK SGEKGINTMD LAQVTGQDPR SVTGRIKKIN HLLTSSQLIY K GHVVKQLK LKKFSHDGVD SNPYINIRDH LATIVEVVKR SKNGIRQIID LKRELKFDKE KRLSKAFIAA IAWLDEKEYL KK VLVVSPK NPAIKIRCVK YVKDIPDSKG SPSFEYDSNS ADEDSVSDSK AAFEDEDLVE GLDNFNATDL LQNQGLVMEE KED AVKNEV LLNRFYPLQN QTYDIADKSG LKGISTMDVV NRITGKEFQR AFTKSSEYYL ESVDKQKENT GGYRLFRIYD FEGK KKFFR LFTAQNFQKL TNAEDEISVP KGFDELGKSR TDLKTLNEDN FVALNNTVRF TTDSDGQDIF FWHGELKIPP NSKKT PNKN KRKRQVKNST NASVAGNISN PKRIKLEQHV STAQEPKSAE DSPSSNGGTV VKGKVVNFGG FSARSLRSLQ RQRAIL KVM NTIGGVAYLR EQFYESVSKY MGSTTTLDKK TVRGDVDLMV ESEKLGARTE PVSGRKIIFL PTVGEDAIQR YILKEKD SK KATFTDVIHD TEIYFFDQTE KNRFHRGKKS VERIRKFQNR QKNAKIKASD DAISKKSTSV NVSDGKIKRR DKKVSAGR T TVVVENTKED KTVYHAGTKD GVQALIRAVV VTKSIKNEIM WDKITKLFPN NSLDNLKKKW TARRVRMGHS GWRAYVDKW KKMLVLAIKS EKISLRDVEE LDLIKLLDIW TSFDEKEIKR PLFLYKNYEE NRKKFTLVRD DTLTHSGNDL AMSSMIQREI SSLKKTYTR KISASTKDLS KSQSDDYIRT VIRSILIESP STTRNEIEAL KNVGNESIDN VIMDMAKEKQ IYLHGSKLEC T DTLPDILE NRGNYKDFGV AFQYRCKVNE LLEAGNAIVI NQEPSDISSW VLIDLISGEL LNMDVIPMVR NVRPLTYTSR RF EIRTLTP PLIIYANSQT KLNTARKSAV KVPLGKPFSR LWVNGSGSIR PNIWKQVVTM VVNEIIFHPG ITLSRLQSRC REV LSLHEI SEICKWLLER QVLITTDFDG YWVNHNWYSI YESTENLYFQ SMSASAWSHP QFEKGGGSGG GSGGSAWSHP QFEK S UniProtKB: Transcription factor tau 138 kDa subunit |
-Macromolecule #2: Transcription factor tau 131 kDa subunit
| Macromolecule | Name: Transcription factor tau 131 kDa subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 120.746969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSERD SSLLAEFSDY GEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR KERVLDPEVA QLLSQANEAF VRNDLQVAER LFNEVIKKDA R NFAAYETL ...String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSERD SSLLAEFSDY GEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR KERVLDPEVA QLLSQANEAF VRNDLQVAER LFNEVIKKDA R NFAAYETL GDIYQLQGRL NDCCNSWFLA AHLNASDWEF WKIVAILSAD LDHVRQAIYC FSRVISLNPM EWESIYRRSM LY KKTGQLA RALDGFQRLY MYNPYDANIL RELAILYVDY DRIEDSIELY MKVFNANVER REAILAALEN ALDSSDEESA AEG EDADEK EPLEQDEDRQ MFPDINWKKI DAKYKCIPFD WSSLNILAEL FLKLAVSEVD GIKTIKKCAR WIQRRESQTF WDHV PDDSE FDNRRFKNST FDSLLAAEKE KSYNIPIDIR VRLGLLRLNT DNLVEALNHF QCLYDETFSD VADLYFEAAT ALTRA EKYK EAIDFFTPLL SLEEWRTTDV FKPLARCYKE IESYETAKEF YELAIKSEPD DLDIRVSLAE VYYRLNDPET FKHMLV DVV EMRKHQVDET LHRISNEKSS NDTSDISSKP LLEDSKFRTF RKKKRTPYDA ERERIERERR ITAKVVDKYE KMKKFEL NS GLNEAKQASI WINTVSELVD IFSSVKNFFM KSRSRKFVGI LRRTKKFNTE LDFQIERLSK LAEGDSVFEG PLMEERVT L TSATELRGLS YEQWFELFME LSLVIAKYQS VEDGLSVVET AQEVNVFFQD PERVKMMKFV KLAIVLQMDD EEELAENLR GLLNQFQFNR KVLQVFMYSL CRGPSSLNIL SSTIQQKFFL RQLKAFDSCR YNTEVNGQAS ITNKEVYNPN KKSSPYLYYI YAVLLYSSR GFLSALQYLT RLEEDIPDDP MVNLLMGLSH IHRAMQRLTA QRHFQIFHGL RYLYRYHKIR KSLYTDLEKQ E ADYNLGRA FHLIGLVSIA IEYYNRVLEN YDDGKLKKHA AYNSIIIYQQ SGNVELADHL MEKYLSIRSG G UniProtKB: Transcription factor tau 131 kDa subunit |
-Macromolecule #3: Transcription factor tau 95 kDa subunit
| Macromolecule | Name: Transcription factor tau 95 kDa subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 69.066438 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHENLY FQSMPVEEPL ATLSSIPDSS ADQAPPLIAD EFTLDLPRIP SLELPLNVST KHSSIQKAIK MCGGIEKVKE AFKEHGPIE SQHGLQLYLN DDTDSDGSKS YFNEHPVIGK RVPFRDESVI LKVTMPKGTL SKNNNSVKDS IKSLKDSNKL R VTPVSIVD ...String: HHHHHHENLY FQSMPVEEPL ATLSSIPDSS ADQAPPLIAD EFTLDLPRIP SLELPLNVST KHSSIQKAIK MCGGIEKVKE AFKEHGPIE SQHGLQLYLN DDTDSDGSKS YFNEHPVIGK RVPFRDESVI LKVTMPKGTL SKNNNSVKDS IKSLKDSNKL R VTPVSIVD NTIKFREMSD FQIKLDNVPS AREFKSSFGS LEWNNFKSFV NSVPDNDSQP QENIGNLILD RSVKIPSTDF QL PPPPKLS MVGFPLLYKY KANPFAKKKK NGVTEVKGTY IKNYQLFVHD LSDKTVIPSQ AHEQVLYDFE VAKKTKVYPG TKS DSKFYE SLEECLKILR ELFARRPIWV KRHLDGIVPK KIHHTMKIAL ALISYRFTMG PWRNTYIKFG IDPRSSVEYA QYQT EYFKI ERKLLSSPIV KKNVPKPPPL VFESDTPGGI DSRFKFDGKR IPWYLMLQID LLIGEPNIAE VFHNVEYLDK ANELT GWFK ELDLVKIRRI VKYELGCMVQ GNYEYNKYKL KYFKTMLFVK ESMVPENKNS EEGMGVNTNK DADGDINMDA GSQMSS NAI EEDKGIAAGD DFDDNGAITE EPDDAALENE EMDTDQNLKV PAS UniProtKB: Transcription factor tau 95 kDa subunit |
-Macromolecule #4: Transcription factor tau 55 kDa subunit
| Macromolecule | Name: Transcription factor tau 55 kDa subunit / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.198898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRCL ETVQPIAKLL EIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI SQEWDSTLTP NEKGETEQEM YMRFKKFWPL FIERVEKEYP N VECILLVT ...String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRCL ETVQPIAKLL EIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI SQEWDSTLTP NEKGETEQEM YMRFKKFWPL FIERVEKEYP N VECILLVT HAASKIALGM SLLGYDNPRM SLNENGDKIR SGSCSLDKYE ILKKSYDTID ETDDQTSFTY IPFSDRKWVL TM NGNTEFL SSGEEMNWNF DCVAEAGSDA DIKKRQMTKK TSSPIPEADD QTEVETVYIS VDIPSGNYKE RTEIAKSAIL QYS GLETDA PLFRIGNRLY EGSWERLVGT ELAFPNAAHV HKKTAGLLSP TEENETTNAG QSKGSSTAND PNIQIQEEDV GLPD STNTS RDHTGDKEEV QSEKIYRIKE RIVLSNVRPM UniProtKB: Transcription factor tau 55 kDa subunit |
-Macromolecule #5: DNA (45-MER)
| Macromolecule | Name: DNA (45-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.643719 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT)(DT)(DT) |
-Macromolecule #6: DNA (45-MER)
| Macromolecule | Name: DNA (45-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.049344 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA)(DA)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 39.6 e/Å2 Details: dataset1: 39.6 dataset2: 43.6 dataset3: 43.2 dataset4: 43.2 dataset5: 44.4 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL / In silico model: Alphafol multimer V2.3.0 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 114621 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)