+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
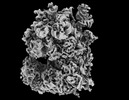
| タイトル | Human NatA-MAP2 80S ribosome complex | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | co-translational processing / ribosome associated factor (RAF) / methionine aminopeptidase 2 (MAP2) / N-terminal methionine excision (NME) / N-acetyl-transferase A (NatA) / N-termional acetylation (NTA) / UBA domain / a-solenoid / protein-protein and protein-RNA interactions / TRANSLATION | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of maintenance of mitotic sister chromatid cohesion, centromeric / protein-N-terminal-glutamate acetyltransferase activity / N-terminal amino-acid Nalpha-acetyltransferase NatA / N-terminal protein amino acid acetylation / NatA complex / protein N-terminal-serine acetyltransferase activity / protein-N-terminal-alanine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / internal protein amino acid acetylation / N-acetyltransferase activity ...negative regulation of maintenance of mitotic sister chromatid cohesion, centromeric / protein-N-terminal-glutamate acetyltransferase activity / N-terminal amino-acid Nalpha-acetyltransferase NatA / N-terminal protein amino acid acetylation / NatA complex / protein N-terminal-serine acetyltransferase activity / protein-N-terminal-alanine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / internal protein amino acid acetylation / N-acetyltransferase activity / methionyl aminopeptidase / initiator methionyl aminopeptidase activity / acetyltransferase activator activity / metalloexopeptidase activity / eukaryotic 80S initiation complex / regulation of translation involved in cellular response to UV / axial mesoderm development / 90S preribosome assembly / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / positive regulation of DNA damage response, signal transduction by p53 class mediator / TORC2 complex binding / regulation of translational initiation / middle ear morphogenesis / protein acetylation / metalloaminopeptidase activity / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / chromosome organization / L13a-mediated translational silencing of Ceruloplasmin expression / aminopeptidase activity / Major pathway of rRNA processing in the nucleolus and cytosol / protein-RNA complex assembly / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / rough endoplasmic reticulum / cytosolic ribosome / ossification / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of translation / ribosomal large subunit biogenesis / skeletal system development / DNA damage response, signal transduction by p53 class mediator / sensory perception of sound / cellular response to gamma radiation / Regulation of expression of SLITs and ROBOs / protein processing / mRNA 5'-UTR binding / cytoplasmic ribonucleoprotein granule / rRNA processing / cellular response to UV / Inactivation, recovery and regulation of the phototransduction cascade / regulation of translation / ribosome binding / cell body / angiogenesis / transcription regulator complex / cytosolic large ribosomal subunit / cytoplasmic translation / cell differentiation / postsynaptic density / protein stabilization / rRNA binding / nuclear body / structural constituent of ribosome / cadherin binding / translation / ribonucleoprotein complex / focal adhesion / mRNA binding / intracellular membrane-bounded organelle / synapse / dendrite / negative regulation of apoptotic process / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / nucleolus / endoplasmic reticulum / DNA binding / RNA binding / extracellular exosome / nucleoplasm / metal ion binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.69 Å | |||||||||
 データ登録者 データ登録者 | Klein MA / Wild K / Sinning I | |||||||||
| 資金援助 |  ドイツ, 1件 ドイツ, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2024 ジャーナル: Nat Commun / 年: 2024タイトル: Multi-protein assemblies orchestrate co-translational enzymatic processing on the human ribosome. 著者: Marius Klein / Klemens Wild / Irmgard Sinning /  要旨: Nascent chains undergo co-translational enzymatic processing as soon as their N-terminus becomes accessible at the ribosomal polypeptide tunnel exit (PTE). In eukaryotes, N-terminal methionine ...Nascent chains undergo co-translational enzymatic processing as soon as their N-terminus becomes accessible at the ribosomal polypeptide tunnel exit (PTE). In eukaryotes, N-terminal methionine excision (NME) by Methionine Aminopeptidases (MAP1 and MAP2), and N-terminal acetylation (NTA) by N-Acetyl-Transferase A (NatA), is the most common combination of subsequent modifications carried out on the 80S ribosome. How these enzymatic processes are coordinated in the context of a rapidly translating ribosome has remained elusive. Here, we report two cryo-EM structures of multi-enzyme complexes assembled on vacant human 80S ribosomes, indicating two routes for NME-NTA. Both assemblies form on the 80S independent of nascent chain substrates. Irrespective of the route, NatA occupies a non-intrusive 'distal' binding site on the ribosome which does not interfere with MAP1 or MAP2 binding nor with most other ribosome-associated factors (RAFs). NatA can partake in a coordinated, dynamic assembly with MAP1 through the hydra-like chaperoning function of the abundant Nascent Polypeptide-Associated Complex (NAC). In contrast to MAP1, MAP2 completely covers the PTE and is thus incompatible with NAC and MAP1 recruitment. Together, our data provide the structural framework for the coordinated orchestration of NME and NTA in protein biogenesis. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_50641.map.gz emd_50641.map.gz | 727.3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-50641-v30.xml emd-50641-v30.xml emd-50641.xml emd-50641.xml | 31.9 KB 31.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_50641_fsc.xml emd_50641_fsc.xml | 23.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_50641.png emd_50641.png | 90.1 KB | ||
| Filedesc metadata |  emd-50641.cif.gz emd-50641.cif.gz | 10.1 KB | ||
| その他 |  emd_50641_half_map_1.map.gz emd_50641_half_map_1.map.gz emd_50641_half_map_2.map.gz emd_50641_half_map_2.map.gz | 1.3 GB 1.3 GB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50641 http://ftp.pdbj.org/pub/emdb/structures/EMD-50641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50641 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_50641_validation.pdf.gz emd_50641_validation.pdf.gz | 1.3 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_50641_full_validation.pdf.gz emd_50641_full_validation.pdf.gz | 1.3 MB | 表示 | |
| XML形式データ |  emd_50641_validation.xml.gz emd_50641_validation.xml.gz | 33.8 KB | 表示 | |
| CIF形式データ |  emd_50641_validation.cif.gz emd_50641_validation.cif.gz | 45.1 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50641 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50641 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  9fpzMC  9fq0C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_50641.map.gz / 形式: CCP4 / 大きさ: 1.4 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_50641.map.gz / 形式: CCP4 / 大きさ: 1.4 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
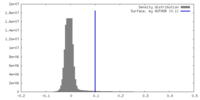
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #1
| ファイル | emd_50641_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
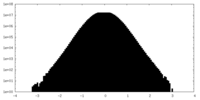
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_50641_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
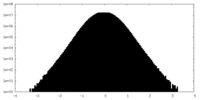
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : human NatA and MAP2 bound to the PTE of the 80S ribosome
+超分子 #1: human NatA and MAP2 bound to the PTE of the 80S ribosome
+分子 #1: N-alpha-acetyltransferase 10
+分子 #3: Methionine aminopeptidase 2
+分子 #4: N-alpha-acetyltransferase 15, NatA auxiliary subunit
+分子 #6: 60S ribosomal protein L4
+分子 #7: Large ribosomal subunit protein eL6
+分子 #8: 60S ribosomal protein L38
+分子 #9: Large ribosomal subunit protein uL24
+分子 #10: 60S ribosomal protein L35
+分子 #11: 60S ribosomal protein L23a
+分子 #12: 60S ribosomal protein L19
+分子 #13: 60S ribosomal protein L28
+分子 #2: 5.8S rRNA (58-MER)
+分子 #5: 28S rRNA
+分子 #14: COBALT (II) ION
+分子 #15: INOSITOL HEXAKISPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 41.28 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.3000000000000003 µm 最小 デフォーカス(公称値): 1.3 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)