+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Corynebacterium glutamicum PS2 S-layer | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | S-layer protein / 2D-lattice / hexagonal lattice / STRUCTURAL PROTEIN | |||||||||||||||||||||
| Function / homology | membrane / PS2 Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) | |||||||||||||||||||||
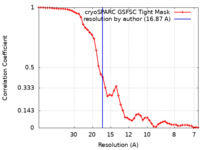
| Method | single particle reconstruction / cryo EM / Resolution: 16.87 Å | |||||||||||||||||||||
 Authors Authors | Isbilir B / Bharat TAM | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  France, European Union, 6 items France, European Union, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Punctuated and continuous structural diversity of S-layers across the prokaryotic tree of life Authors: Johnston E / Isbilir B / Alva V / Bharat TAM / Doye JPK | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50640.map.gz emd_50640.map.gz | 6.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50640-v30.xml emd-50640-v30.xml emd-50640.xml emd-50640.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50640_fsc.xml emd_50640_fsc.xml | 4.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_50640.png emd_50640.png | 64.3 KB | ||
| Filedesc metadata |  emd-50640.cif.gz emd-50640.cif.gz | 5.7 KB | ||
| Others |  emd_50640_additional_1.map.gz emd_50640_additional_1.map.gz emd_50640_half_map_1.map.gz emd_50640_half_map_1.map.gz emd_50640_half_map_2.map.gz emd_50640_half_map_2.map.gz | 12.1 MB 11.8 MB 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50640 http://ftp.pdbj.org/pub/emdb/structures/EMD-50640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50640 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50640.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50640.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.296 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map
| File | emd_50640_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_50640_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_50640_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of Hexagonally Packed PS2 Protein
| Entire | Name: Structure of Hexagonally Packed PS2 Protein |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Hexagonally Packed PS2 Protein
| Supramolecule | Name: Structure of Hexagonally Packed PS2 Protein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) / Strain: ATCC13058 / Location in cell: extracellular Corynebacterium glutamicum (bacteria) / Strain: ATCC13058 / Location in cell: extracellular |
| Molecular weight | Theoretical: 54 KDa |
-Macromolecule #1: PS2 - Corynebacterium glutamicum S-layer protein
| Macromolecule | Name: PS2 - Corynebacterium glutamicum S-layer protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Corynebacterium glutamicum (bacteria) Corynebacterium glutamicum (bacteria) |
| Sequence | String: MFNNRIRTAA LAGAIAISTA ASGVAIPAFA QETGTYNNTG GFNDADGSTI QPAPAVDHSE AELRDATDA TGNYLAAFQS GDIEAIVGAY IDVGVDGFDP SEEAIFKAFE AARDEATQQL A FSAETITK TRESVAYALK VDQEATEAYL AYRNALRGAA TSINPLIDAA ...String: MFNNRIRTAA LAGAIAISTA ASGVAIPAFA QETGTYNNTG GFNDADGSTI QPAPAVDHSE AELRDATDA TGNYLAAFQS GDIEAIVGAY IDVGVDGFDP SEEAIFKAFE AARDEATQQL A FSAETITK TRESVAYALK VDQEATEAYL AYRNALRGAA TSINPLIDAA NAANRTDGSE IE IYDNIFL ASDVFTDGPL LLPAYRELVA LQTEVNEDLE WLGEFAIDND ADNYVQRYHI PAV EALKAE IDARLEAIEP HRADSAEKNR LAQKSDVLVR QLFLERATAQ RDTLRIVEAI FATA TRYVE LYESDENINV ENKTLREHYF ALFPTLFGAA SFNVGVLNTA DDAAIDYYLV WDTDL ETND EDAAYAEEKR EFALLTYAKI FINGQWQEKV KYVQNLDDGA RAEAARIEAE RLADEA YRA EQLRIAQEAA DAQKAIADAL AKEAENNNNS GGDNSSDDKG TGSSDIGSWG PFAAIAA II AAIAAIFPFL SGIVKF UniProtKB: PS2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 50 mM HEPES pH=7.5, 150 mM NaCl, 1 mM MgCl2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: Absorption for 20 sec and blotted for 2.5 sec with blot force -5.. | ||||||||||||
| Details | Purified Surface Layer of Corynebacterium glutamicum composed of PS2 protein |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 31 / Average exposure time: 3.28 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 96000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)