[English] 日本語
 Yorodumi
Yorodumi- EMDB-50621: Structure of heteromeric amyloid filament of TDP-43 and AXNA11 fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 2) | |||||||||
 Map data Map data | TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TDP-43 / ANXA11 / amyloid / heteromeric amyloid / FTLD-TDP / neurodegeneration / dementia / brain / protein filament / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytokinetic process / nuclear inner membrane organization / interchromatin granule / perichromatin fibrils / 3'-UTR-mediated mRNA destabilization / specific granule / calcium-dependent phospholipid binding / phosphatidylethanolamine binding / S100 protein binding / vesicle membrane ...cytokinetic process / nuclear inner membrane organization / interchromatin granule / perichromatin fibrils / 3'-UTR-mediated mRNA destabilization / specific granule / calcium-dependent phospholipid binding / phosphatidylethanolamine binding / S100 protein binding / vesicle membrane / azurophil granule / 3'-UTR-mediated mRNA stabilization / intracellular membraneless organelle / phosphatidylserine binding / negative regulation of protein phosphorylation / host-mediated suppression of viral transcription / phagocytosis / pre-mRNA intronic binding / phagocytic vesicle / RNA splicing / response to endoplasmic reticulum stress / mRNA 3'-UTR binding / molecular condensate scaffold activity / response to calcium ion / regulation of circadian rhythm / positive regulation of insulin secretion / regulation of protein stability / positive regulation of protein import into nucleus / spindle / mRNA processing / cytoplasmic stress granule / calcium-dependent protein binding / MHC class II protein complex binding / rhythmic process / melanosome / nuclear envelope / regulation of gene expression / : / double-stranded DNA binding / midbody / regulation of apoptotic process / amyloid fibril formation / regulation of cell cycle / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / negative regulation of gene expression / intracellular membrane-bounded organelle / lipid binding / calcium ion binding / chromatin / mitochondrion / DNA binding / RNA binding / extracellular exosome / nucleoplasm / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Arseni D / Ryskeldi-Falcon B | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Heteromeric amyloid filaments of ANXA11 and TDP-43 in FTLD-TDP Type C. Authors: Diana Arseni / Takashi Nonaka / Max H Jacobsen / Alexey G Murzin / Laura Cracco / Sew Y Peak-Chew / Holly J Garringer / Ito Kawakami / Hisaomi Suzuki / Misumoto Onaya / Yuko Saito / Shigeo ...Authors: Diana Arseni / Takashi Nonaka / Max H Jacobsen / Alexey G Murzin / Laura Cracco / Sew Y Peak-Chew / Holly J Garringer / Ito Kawakami / Hisaomi Suzuki / Misumoto Onaya / Yuko Saito / Shigeo Murayama / Changiz Geula / Ruben Vidal / Kathy L Newell / Marsel Mesulam / Bernardino Ghetti / Masato Hasegawa / Benjamin Ryskeldi-Falcon Abstract: Neurodegenerative diseases are characterised by the abnormal filamentous assembly of specific proteins in the central nervous system . Human genetic studies established a causal role for protein ...Neurodegenerative diseases are characterised by the abnormal filamentous assembly of specific proteins in the central nervous system . Human genetic studies established a causal role for protein assembly in neurodegeneration . However, the underlying molecular mechanisms remain largely unknown, which is limiting progress in developing clinical tools for these diseases. Recent advances in electron cryo-microscopy (cryo-EM) have enabled the structures of the protein filaments to be determined from patient brains . All diseases studied to date have been characterised by the self-assembly of a single intracellular protein in homomeric amyloid filaments, including that of TAR DNA-binding protein 43 (TDP-43) in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) Types A and B . Here, we used cryo-EM to determine filament structures from the brains of individuals with FTLD-TDP Type C, one of the most common forms of sporadic FTLD-TDP. Unexpectedly, the structures revealed that a second protein, annexin A11 (ANXA11), co-assembles with TDP-43 in heteromeric amyloid filaments. The ordered filament fold is formed by TDP-43 residues G282/284-N345 and ANXA11 residues L39-L74 from their respective low-complexity domains (LCDs). Regions of TDP-43 and ANXA11 previously implicated in protein-protein interactions form an extensive hydrophobic interface at the centre of the filament fold. Immunoblots of the filaments revealed that the majority of ANXA11 exists as a ∼22 kDa N-terminal fragment (NTF) lacking the annexin core domain. Immunohistochemistry of brain sections confirmed the co-localisation of ANXA11 and TDP-43 in inclusions, redefining the histopathology of FTLD-TDP Type C. This work establishes a central role for ANXA11 in FTLD-TDP Type C. The unprecedented formation of heteromeric amyloid filaments in human brain revises our understanding of amyloid assembly and may be of significance for the pathogenesis of neurodegenerative diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50621.map.gz emd_50621.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50621-v30.xml emd-50621-v30.xml emd-50621.xml emd-50621.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50621.png emd_50621.png | 52.7 KB | ||
| Masks |  emd_50621_msk_1.map emd_50621_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50621.cif.gz emd-50621.cif.gz | 5.4 KB | ||
| Others |  emd_50621_half_map_1.map.gz emd_50621_half_map_1.map.gz emd_50621_half_map_2.map.gz emd_50621_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50621 http://ftp.pdbj.org/pub/emdb/structures/EMD-50621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50621 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50621 | HTTPS FTP |
-Validation report
| Summary document |  emd_50621_validation.pdf.gz emd_50621_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50621_full_validation.pdf.gz emd_50621_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_50621_validation.xml.gz emd_50621_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  emd_50621_validation.cif.gz emd_50621_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50621 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50621 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50621 | HTTPS FTP |
-Related structure data
| Related structure data |  9fofMC  9forC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50621.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50621.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
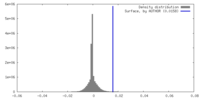
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50621_msk_1.map emd_50621_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
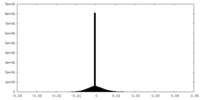
| Projections & Slices |
| ||||||||||||
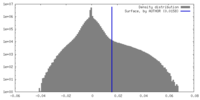
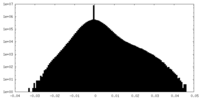
| Density Histograms |
-Half map: TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 half 1
| File | emd_50621_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 half 1 | ||||||||||||
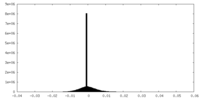
| Projections & Slices |
| ||||||||||||
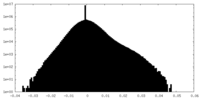
| Density Histograms |
-Half map: TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 half 2
| File | emd_50621_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TDP-43/ANXA11 FTLD-TDP type C heteroamyloid filament variant 2 half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP T...
| Entire | Name: heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 1) |
|---|---|
| Components |
|
-Supramolecule #1: heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP T...
| Supramolecule | Name: heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 1) type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Brain Homo sapiens (human) / Tissue: Brain |
-Macromolecule #1: TAR DNA-binding protein 43
| Macromolecule | Name: TAR DNA-binding protein 43 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Brain Homo sapiens (human) / Tissue: Brain |
| Molecular weight | Theoretical: 6.23187 KDa |
| Sequence | String: GFGNQGGFGN SRGGGAGLGN NQGSNMGGGM NFGAFSINPA MMAAAQAALQ SSWGMMGMLA SQQN UniProtKB: TAR DNA-binding protein 43 |
-Macromolecule #2: Annexin A11
| Macromolecule | Name: Annexin A11 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Brain Homo sapiens (human) / Tissue: Brain |
| Molecular weight | Theoretical: 3.805148 KDa |
| Sequence | String: LDNVATYAGQ FNQDYLSGMA ANMSGTFGGA NMPNLY UniProtKB: Annexin A11 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 4.96 Å Applied symmetry - Helical parameters - Δ&Phi: -1.92 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 10842 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)