[English] 日本語
 Yorodumi
Yorodumi- EMDB-50358: In vitro-induced genome-releasing intermediate of Rhodobacter mic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry | ||||||||||||
 Map data Map data | Final run_class001.mrc output from RELION 3.1 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ssDNA virus / Microviridae / Tainavirinae / Alphaproteobacteria / Rhodobacter capsulatus / aquatic virus / jelly-roll fold / VIRUS | ||||||||||||
| Biological species |  Rhodobacter capsulatus (bacteria) / Rhodobacter capsulatus (bacteria) /  Rhodobacter phage Ebor (virus) Rhodobacter phage Ebor (virus) | ||||||||||||
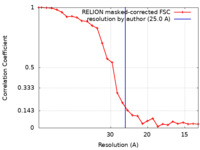
| Method | single particle reconstruction / cryo EM / Resolution: 25.0 Å | ||||||||||||
 Authors Authors | Bardy P / MacDonald CIW / Jenkins HT / Chechik M / Hart SJ / Turkenburg JP / Blaza JN / Fogg PCM / Antson AA | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: A stargate mechanism of genome delivery unveiled by cryogenic electron tomography. Authors: Pavol Bardy / Conor I W MacDonald / Paul C Kirchberger / Huw T Jenkins / Tibor Botka / Lewis Byrom / Nawshin T B Alim / Daouda A K Traore / Hannah C König / Tristan R Nicholas / Maria ...Authors: Pavol Bardy / Conor I W MacDonald / Paul C Kirchberger / Huw T Jenkins / Tibor Botka / Lewis Byrom / Nawshin T B Alim / Daouda A K Traore / Hannah C König / Tristan R Nicholas / Maria Chechik / Samuel J Hart / Johan P Turkenburg / James N Blaza / J Thomas Beatty / Paul C M Fogg / Alfred A Antson /     Abstract: Single-stranded DNA bacteriophages of the family are major components of the global virosphere. Microviruses are highly abundant in aquatic ecosystems and are prominent members of the mammalian gut ...Single-stranded DNA bacteriophages of the family are major components of the global virosphere. Microviruses are highly abundant in aquatic ecosystems and are prominent members of the mammalian gut microbiome, where their diversity has been linked to various chronic health disorders. Despite the clear importance of microviruses, little is known about the molecular mechanism of host infection. Here, we have characterized an exceptionally large microvirus, Ebor, and provide crucial insights into long-standing mechanistic questions. Cryogenic electron microscopy of Ebor revealed a capsid with trimeric protrusions that recognise lipopolysaccharides on the host surface. Cryogenic electron tomography of the host cell colonized with virus particles demonstrated that the virus initially attaches to the cell via five such protrusions, located at the corners of a single pentamer. This interaction triggers a stargate mechanism of capsid opening along the 5-fold symmetry axis, enabling delivery of the virus genome. Despite variations in specific virus-host interactions among different family viruses, structural data indicate that the stargate mechanism of infection is universally employed by all members of the family. Startlingly, our data reveal a mechanistic link for the opening of relatively small capsids made out of a single jelly-roll fold with the structurally unrelated giant viruses. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50358.map.gz emd_50358.map.gz | 721.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50358-v30.xml emd-50358-v30.xml emd-50358.xml emd-50358.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50358_fsc.xml emd_50358_fsc.xml | 2.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_50358.png emd_50358.png | 27 KB | ||
| Filedesc metadata |  emd-50358.cif.gz emd-50358.cif.gz | 5.9 KB | ||
| Others |  emd_50358_half_map_1.map.gz emd_50358_half_map_1.map.gz emd_50358_half_map_2.map.gz emd_50358_half_map_2.map.gz | 732 KB 731.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50358 http://ftp.pdbj.org/pub/emdb/structures/EMD-50358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50358 | HTTPS FTP |
-Validation report
| Summary document |  emd_50358_validation.pdf.gz emd_50358_validation.pdf.gz | 708.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50358_full_validation.pdf.gz emd_50358_full_validation.pdf.gz | 708.4 KB | Display | |
| Data in XML |  emd_50358_validation.xml.gz emd_50358_validation.xml.gz | 7.8 KB | Display | |
| Data in CIF |  emd_50358_validation.cif.gz emd_50358_validation.cif.gz | 9.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50358 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50358 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50358 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50358.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50358.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final run_class001.mrc output from RELION 3.1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.75 Å | ||||||||||||||||||||||||||||||||||||
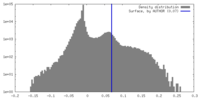
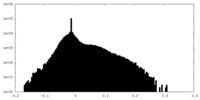
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_50358_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
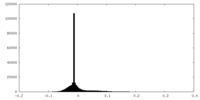
| Density Histograms |
-Half map: #1
| File | emd_50358_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
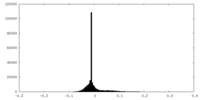
| Density Histograms |
- Sample components
Sample components
-Entire : Rhodobacter phage Ebor
| Entire | Name:  Rhodobacter phage Ebor (virus) Rhodobacter phage Ebor (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Rhodobacter phage Ebor
| Supramolecule | Name: Rhodobacter phage Ebor / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: purified from Rhodobacter capsulatus strain B10 by CsCl ultracentrifugation NCBI-ID: 3144506 / Sci species name: Rhodobacter phage Ebor / Sci species strain: variant S120 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Rhodobacter capsulatus (bacteria) / Strain: B10 Rhodobacter capsulatus (bacteria) / Strain: B10 |
| Molecular weight | Theoretical: 5.54 MDa |
| Virus shell | Shell ID: 1 / Name: Genome-releasing capsid / Diameter: 280.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus (bacteria) / Strain: B10 Rhodobacter capsulatus (bacteria) / Strain: B10 |
| Sequence | String: MSYQTKRNVR REARTLMGRF KAGKLAPVMA VPVKGSEGGM LSQSVSFELD PIAGRMATPI TAEMCAVFVP VQACDALKNP EADYAGMTEI VREKLLSGNP LFVLEPETDV SKRCGVNPRS NNGLMRVNEI VRLAHNCAVN FLRRRRYVDA VQLTAANHST TPAILSQTVL ...String: MSYQTKRNVR REARTLMGRF KAGKLAPVMA VPVKGSEGGM LSQSVSFELD PIAGRMATPI TAEMCAVFVP VQACDALKNP EADYAGMTEI VREKLLSGNP LFVLEPETDV SKRCGVNPRS NNGLMRVNEI VRLAHNCAVN FLRRRRYVDA VQLTAANHST TPAILSQTVL DRFNGALDPD PNVNGAVQLS MPDMRLPVAS DAEKAVTAPL TVKRGNENRR LDAGISRLAA AEVAAATDAA LYAVFGGREA GNVSLTDFYN AQKMDELTRV MRKICDDNPE YGEEMVLRWA HGLSVDPGRV PFLLAEKSVV LGRQIIGATD TAGVEDGVKR SDMAAQLSFT VPIPTTELGG IIVTFACIKP DETLSSQPHP ILADHWRLDN FVADELALDP QPVMARELDY KVAQANETTV VFYTGLNELK KTYVSYGLCR ALDPNTVESK NAVWQLEVPL SVTPETVLYP ADLPQYPFAD QQAEVCTYVV QSTAVMPTPM IFGPSPVEQL AVIETEDLFE D |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.75 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot time 2 sec, blot force 0, wait time 8 sec. | ||||||||||||
| Details | concentration of 3x10^10 PFU/ml |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 2810 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)