[English] 日本語
 Yorodumi
Yorodumi- EMDB-50341: Cryo-EM structure of the beta3 homomeric GABA(A) receptor in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the beta3 homomeric GABA(A) receptor in complex with HSM in the long-lived symmetric desensitised state | |||||||||||||||||||||
 Map data Map data | Unsharpened map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | GABA / neurotransmission / gating cycle / time-resolved cryo-EM / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcircadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex ...circadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / response to anesthetic / postsynaptic specialization membrane / inhibitory postsynaptic potential / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / cellular response to zinc ion / exploration behavior / motor behavior / roof of mouth development / Signaling by ERBB4 / cochlea development / social behavior / chloride channel complex / chloride transmembrane transport / cytoplasmic vesicle membrane / cerebellum development / learning / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / memory / dendritic spine / postsynaptic membrane / response to xenobiotic stimulus / cell surface / signal transduction / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||||||||||||||
 Authors Authors | Mihaylov DB / Malinauskas T / Aricescu AR | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the beta3 homomeric GABA(A) receptor in complex with HSM in the long-lived symmetric desensitised state Authors: Mihaylov DB / Malinauskas T / Aricescu AR | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50341.map.gz emd_50341.map.gz | 93.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50341-v30.xml emd-50341-v30.xml emd-50341.xml emd-50341.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50341_fsc.xml emd_50341_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_50341.png emd_50341.png | 99 KB | ||
| Masks |  emd_50341_msk_1.map emd_50341_msk_1.map | 120.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50341.cif.gz emd-50341.cif.gz | 7.1 KB | ||
| Others |  emd_50341_additional_1.map.gz emd_50341_additional_1.map.gz emd_50341_half_map_1.map.gz emd_50341_half_map_1.map.gz emd_50341_half_map_2.map.gz emd_50341_half_map_2.map.gz | 11.7 MB 94.7 MB 94.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50341 http://ftp.pdbj.org/pub/emdb/structures/EMD-50341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50341 | HTTPS FTP |
-Related structure data
| Related structure data |  9feuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50341.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50341.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50341_msk_1.map emd_50341_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map
| File | emd_50341_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps
| File | emd_50341_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps
| File | emd_50341_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the beta3 homomeric GABA(A) receptor in comp...
| Entire | Name: Cryo-EM structure of the beta3 homomeric GABA(A) receptor in complex with HSM in the long-lived symmetric desensitised state |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the beta3 homomeric GABA(A) receptor in comp...
| Supramolecule | Name: Cryo-EM structure of the beta3 homomeric GABA(A) receptor in complex with HSM in the long-lived symmetric desensitised state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Gamma-aminobutyric acid receptor subunit beta-3
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit beta-3 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.643246 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPDY DIPTTENLYF QGTGQSVNDP GNMSFVKETV DKLLKGYDIR LRPDFGGPP VCVGMNIDIA SIDMVSEVNM DYTLTMYFQQ YWRDKRLAYS GIPLNLTLDN RVADQLWVPD TYFLNDKKSF V HGVTVKNR ...String: MDEKTTGWRG GHVVEGLAGE LEQLRARLEH HPQGQREPDY DIPTTENLYF QGTGQSVNDP GNMSFVKETV DKLLKGYDIR LRPDFGGPP VCVGMNIDIA SIDMVSEVNM DYTLTMYFQQ YWRDKRLAYS GIPLNLTLDN RVADQLWVPD TYFLNDKKSF V HGVTVKNR MIRLHPDGTV LYGLRITTTA ACMMDLRRYP LDEQNCTLEI ESYGYTTDDI EFYWRGGDKA VTGVERIELP QF SIVEHRL VSRNVVFATG AYPRLSLSFR LKRNIGYFIL QTYMPSILIT ILSWVSFWIN YDASAARVAL GITTVLTMTT INT HLRETL PKIPYVKAID MYLMGCFVFV FLALLEYAFV NYIFFSQPAR AAAIDRWSRI VFPFTFSLFN LVYWLYYVN UniProtKB: Gamma-aminobutyric acid receptor subunit beta-3, Gamma-aminobutyric acid receptor subunit beta-3 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: HISTAMINE
| Macromolecule | Name: HISTAMINE / type: ligand / ID: 5 / Number of copies: 5 / Formula: HSM |
|---|---|
| Molecular weight | Theoretical: 111.145 Da |
| Chemical component information | 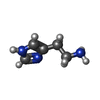 ChemComp-HSM: |
-Macromolecule #6: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 6 / Number of copies: 15 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #7: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 7 / Number of copies: 5 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 370 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO | ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa / Details: current: 30mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 20757 / Average exposure time: 6.43 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 96000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 30 / Target criteria: FSC at 0.5 |
| Output model |  PDB-9feu: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















































