[English] 日本語
 Yorodumi
Yorodumi- EMDB-50317: USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (ordered s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (ordered subset, focused refinement) | |||||||||
 Map data Map data | Globally sharpened map (B-factor 81.5) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | INHIBITOR / DEUBIQUITINASE / COMPLEX / ENZYME-SUBSTRATE / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of error-prone translesion synthesis / monoubiquitinated protein deubiquitination / protein deubiquitination / regulation of DNA repair / response to UV / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants ...positive regulation of error-prone translesion synthesis / monoubiquitinated protein deubiquitination / protein deubiquitination / regulation of DNA repair / response to UV / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of pyruvate metabolism / TRAF6-mediated induction of TAK1 complex within TLR4 complex / NF-kB is activated and signals survival / Regulation of innate immune responses to cytosolic DNA / Pexophagy / Downregulation of ERBB2:ERBB3 signaling / NRIF signals cell death from the nucleus / Activated NOTCH1 Transmits Signal to the Nucleus / Regulation of PTEN localization / VLDLR internalisation and degradation / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Translesion synthesis by REV1 / Regulation of BACH1 activity / TICAM1, RIP1-mediated IKK complex recruitment / MAP3K8 (TPL2)-dependent MAPK1/3 activation / Translesion synthesis by POLK / InlB-mediated entry of Listeria monocytogenes into host cell / Degradation of CDH1 / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Josephin domain DUBs / Downregulation of TGF-beta receptor signaling / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / Degradation of CRY and PER proteins / IKK complex recruitment mediated by RIP1 / Regulation of activated PAK-2p34 by proteasome mediated degradation / PINK1-PRKN Mediated Mitophagy / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / skeletal system development / Autodegradation of Cdh1 by Cdh1:APC/C / TCF dependent signaling in response to WNT / APC/C:Cdc20 mediated degradation of Securin / Regulation of NF-kappa B signaling / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / positive regulation of receptor signaling pathway via JAK-STAT / activated TAK1 mediates p38 MAPK activation / Asymmetric localization of PCP proteins / Ubiquitin-dependent degradation of Cyclin D / SCF-beta-TrCP mediated degradation of Emi1 / NIK-->noncanonical NF-kB signaling / TNFR2 non-canonical NF-kB pathway / Regulation of signaling by CBL / AUF1 (hnRNP D0) binds and destabilizes mRNA / NOTCH3 Activation and Transmission of Signal to the Nucleus / Negative regulators of DDX58/IFIH1 signaling / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / Negative regulation of FGFR3 signaling / Fanconi Anemia Pathway / Deactivation of the beta-catenin transactivating complex / Peroxisomal protein import / Degradation of DVL / Stabilization of p53 / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Negative regulation of FGFR2 signaling / Dectin-1 mediated noncanonical NF-kB signaling / Negative regulation of FGFR4 signaling / Downregulation of SMAD2/3:SMAD4 transcriptional activity / Negative regulation of FGFR1 signaling / Degradation of AXIN / Regulation of TNFR1 signaling / Termination of translesion DNA synthesis / EGFR downregulation / Hh mutants are degraded by ERAD Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
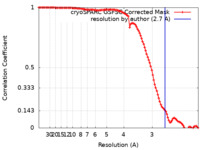
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Rennie ML / Gundogdu M / Walden H | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2024 Journal: J Med Chem / Year: 2024Title: Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279. Authors: Martin Luke Rennie / Mehmet Gundogdu / Connor Arkinson / Steven Liness / Sheelagh Frame / Helen Walden /  Abstract: DNA damage triggers cell signaling cascades that mediate repair. This signaling is frequently dysregulated in cancers. The proteins that mediate this signaling are potential targets for therapeutic ...DNA damage triggers cell signaling cascades that mediate repair. This signaling is frequently dysregulated in cancers. The proteins that mediate this signaling are potential targets for therapeutic intervention. Ubiquitin-specific protease 1 (USP1) is one such target, with small-molecule inhibitors already in clinical trials. Here, we use biochemical assays and cryo-electron microscopy (cryo-EM) to study the clinical USP1 inhibitor, KSQ-4279 (RO7623066), and compare this to the well-established tool compound, ML323. We find that KSQ-4279 binds to the same cryptic site of USP1 as ML323 but disrupts the protein structure in subtly different ways. Inhibitor binding drives a substantial increase in thermal stability of USP1, which may be mediated through the inhibitors filling a hydrophobic tunnel-like pocket in USP1. Our results contribute to the understanding of the mechanism of action of USP1 inhibitors at the molecular level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50317.map.gz emd_50317.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50317-v30.xml emd-50317-v30.xml emd-50317.xml emd-50317.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50317_fsc.xml emd_50317_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_50317.png emd_50317.png | 89.1 KB | ||
| Masks |  emd_50317_msk_1.map emd_50317_msk_1.map emd_50317_msk_2.map emd_50317_msk_2.map emd_50317_msk_3.map emd_50317_msk_3.map | 125 MB 125 MB 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50317.cif.gz emd-50317.cif.gz | 6.9 KB | ||
| Others |  emd_50317_half_map_1.map.gz emd_50317_half_map_1.map.gz emd_50317_half_map_2.map.gz emd_50317_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50317 http://ftp.pdbj.org/pub/emdb/structures/EMD-50317 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50317 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50317 | HTTPS FTP |
-Related structure data
| Related structure data |  9fcjMC  9fciC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50317.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50317.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Globally sharpened map (B-factor 81.5) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
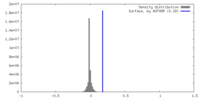
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50317_msk_1.map emd_50317_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
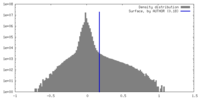
| Density Histograms |
-Mask #2
| File |  emd_50317_msk_2.map emd_50317_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
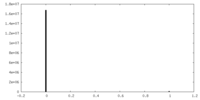
| Density Histograms |
-Mask #3
| File |  emd_50317_msk_3.map emd_50317_msk_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
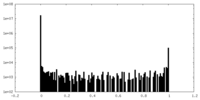
| Density Histograms |
- Sample components
Sample components
-Entire : USP1 BOUND TO ML323 AND UBIQUITIN CONJUGATED TO FANCD2
| Entire | Name: USP1 BOUND TO ML323 AND UBIQUITIN CONJUGATED TO FANCD2 |
|---|---|
| Components |
|
-Supramolecule #1: USP1 BOUND TO ML323 AND UBIQUITIN CONJUGATED TO FANCD2
| Supramolecule | Name: USP1 BOUND TO ML323 AND UBIQUITIN CONJUGATED TO FANCD2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Polyubiquitin-C
| Macromolecule | Name: Polyubiquitin-C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.875125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMQIFVK TLTGKTITLE VEPSDTIENV KAKIQDKEGI PPDQQRLIFA GKQLEDGRTL SDYNIQKEST LHLVLRLRGG UniProtKB: Polyubiquitin-C |
-Macromolecule #2: Ubiquitin carboxyl-terminal hydrolase 1
| Macromolecule | Name: Ubiquitin carboxyl-terminal hydrolase 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: ubiquitinyl hydrolase 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.390273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GMPGVIPSES NGLSRGSPSK KNRLSLKFFQ KKETKRALDF TDSQENEEKA SEYRASEIDQ VVPAAQSSPI NCEKRENLLP FVGLNNLGN TSYLNSILQV LYFCPGFKSG VKHLFNIISR KKEALKDEAN QKDKGNCKED SLASYELICS LQSLIISVEQ L QASFLLNP ...String: GMPGVIPSES NGLSRGSPSK KNRLSLKFFQ KKETKRALDF TDSQENEEKA SEYRASEIDQ VVPAAQSSPI NCEKRENLLP FVGLNNLGN TSYLNSILQV LYFCPGFKSG VKHLFNIISR KKEALKDEAN QKDKGNCKED SLASYELICS LQSLIISVEQ L QASFLLNP EKYTDELATQ PRRLLNTLRE LNPMYEGYLQ HDAQEVLQCI LGNIQETCQL LKKEEVKNVA ELPTKVEEIP HP KEEMNGI NSIEMDSMRH SEDFKEKLPK GNGKRKSDTE FGNMKKKVKL SKEHQSLEEN QRQTRSKRKA TSDTLESPPK IIP KYISEN ESPRPSQKKS RVKINWLKSA TKQPSILSKF CSLGKITTNQ GVKGQSKENE CDPEEDLGKC ESDNTTNGCG LESP GNTVT PVNVNEVKPI NKGEEQIGFE LVEKLFQGQL VLRTRCLECE SLTERREDFQ DISVPVQEDE LSKVEESSEI SPEPK TEMK TLRWAISQFA SVERIVGEDK YFCENCHHYT EAERSLLFDK MPEVITIHLK CFAASGLEFD CYGGGLSKIN TPLLTP LKL SLEEWSTKPT NDSYGLFAVV MHSGITISSG HYTASVKVTD LNSLELDKGN FVVDQMCEIG KPEPLNEEEA RGVVENY ND EEVSIRVGGN TQPSKVLNKK NVEAIGLLAA QKSKADYELY NKASNPDKVA STAFAENRNS ETSDTTGTHE SDRNKESS D QTGINISGFE NKISYVVQSL KEYEGKWLLF DDSEVKVTEE KDFLNSLSPS TSPTSTPYLL FYKKL UniProtKB: Ubiquitin carboxyl-terminal hydrolase 1 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phe...
| Macromolecule | Name: 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine type: ligand / ID: 4 / Number of copies: 1 / Formula: JDA |
|---|---|
| Molecular weight | Theoretical: 384.477 Da |
| Chemical component information | 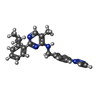 ChemComp-JDA: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 29 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV / Details: blotting for 3.0 seconds. |
| Details | 9.3 uM USP1-UAF1, 1.8 uM FANCI-FANCD2Ub, 2.2 uM dsDNA (61 base-pairs), 18 uM ML323 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)