+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | EVA71 E096A native particle | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | native provirion intermediate / VIRUS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||||||||
| Biological species |   Enterovirus A71 Enterovirus A71 | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||
 Authors Authors | Kingston NJ / Stonehouse NJ / Rowlands DJ / Hogle JM / Filman DJ / Snowden JSS | ||||||||||||
| Funding support |  United States, United States,  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Mechanism of enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate. Authors: Natalie J Kingston / Joseph S Snowden / Keith Grehan / Philippa K Hall / Eero V Hietanen / Tim C Passchier / Stephen J Polyak / David J Filman / James M Hogle / David J Rowlands / Nicola J Stonehouse /   Abstract: Molecular details of genome packaging are little understood for the majority of viruses. In enteroviruses (EVs), cleavage of the structural protein VP0 into VP4 and VP2 is initiated by the ...Molecular details of genome packaging are little understood for the majority of viruses. In enteroviruses (EVs), cleavage of the structural protein VP0 into VP4 and VP2 is initiated by the incorporation of RNA into the assembling virion and is essential for infectivity. We have applied a combination of bioinformatic, molecular and structural approaches to generate the first high-resolution structure of an intermediate in the assembly pathway, termed a provirion, which contains RNA and intact VP0. We have demonstrated an essential role of VP0 E096 in VP0 cleavage independent of RNA encapsidation and generated a new model of capsid maturation, supported by bioinformatic analysis. This provides a molecular basis for RNA-dependence, where RNA induces conformational changes required for VP0 maturation, but that RNA packaging itself is not sufficient to induce maturation. These data have implications for understanding production of infectious virions and potential relevance for future vaccine and antiviral drug design. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50200.map.gz emd_50200.map.gz | 168.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50200-v30.xml emd-50200-v30.xml emd-50200.xml emd-50200.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50200.png emd_50200.png | 129.2 KB | ||
| Masks |  emd_50200_msk_1.map emd_50200_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50200.cif.gz emd-50200.cif.gz | 6.1 KB | ||
| Others |  emd_50200_half_map_1.map.gz emd_50200_half_map_1.map.gz emd_50200_half_map_2.map.gz emd_50200_half_map_2.map.gz | 225.5 MB 225.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50200 http://ftp.pdbj.org/pub/emdb/structures/EMD-50200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50200 | HTTPS FTP |
-Related structure data
| Related structure data |  9f5sM M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50200.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50200.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
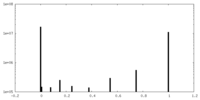
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
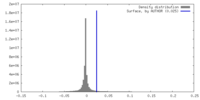
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50200_msk_1.map emd_50200_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
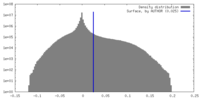
| Density Histograms |
- Sample components
Sample components
-Entire : Enterovirus A71
| Entire | Name:   Enterovirus A71 Enterovirus A71 |
|---|---|
| Components |
|
-Supramolecule #1: Enterovirus A71
| Supramolecule | Name: Enterovirus A71 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Mutant virus recovered from in vitro transcribed RNA NCBI-ID: 39054 / Sci species name: Enterovirus A71 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: VP0
| Macromolecule | Name: VP0 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 35.19523 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSQVSTQRS GSHENSNSAT EGSTINYTTI NYYKDSYAAT AGKQSLKQDP DKFANPVKDV FTEMAAPLKS PSAEACGYSD RVAQLTIGN STITTQAAAN IIVGYGEWPS YCSDDDATAV DKPTRPDVSV NRFYTLDTKL WEKSSKGWYW KFPDVLTETG V FGQNAQFH ...String: MGSQVSTQRS GSHENSNSAT EGSTINYTTI NYYKDSYAAT AGKQSLKQDP DKFANPVKDV FTEMAAPLKS PSAEACGYSD RVAQLTIGN STITTQAAAN IIVGYGEWPS YCSDDDATAV DKPTRPDVSV NRFYTLDTKL WEKSSKGWYW KFPDVLTETG V FGQNAQFH YLYRSGFCIH VQCNASKFHQ GALLVAILPE YVIGTVAGGT GTEDSHPPYI QTQPGADGFE LQHPYVLDAG IP ISQLTVC PHQWINLRTN NCATIIVPYM NTLPFDSALN HCNFGLLVVP ISPLDFDQGA TPVIPITITL APMCSEFAGL RQA VTQ UniProtKB: Genome polyprotein |
-Macromolecule #2: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 26.540332 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRDGPWQST MLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV ...String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRDGPWQST MLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV IPWISNTHYR AHARDGVFDY YTTGLVSIWY QTNYVVPIGA PNTAYIIALA AAQKNFTMKL CKDTSHILQT AS IQ UniProtKB: Genome polyprotein |
-Macromolecule #3: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 32.781805 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GDRVADMIES SIGNSVSRAL TQALPAPTGQ NTQVSSHRLD TGEVPALQAA EIGASSNTSD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLEG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGQVVPQ LLQYMFVPPG A PKPESRES ...String: GDRVADMIES SIGNSVSRAL TQALPAPTGQ NTQVSSHRLD TGEVPALQAA EIGASSNTSD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLEG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGQVVPQ LLQYMFVPPG A PKPESRES LAWQTATNPS VFVKLTDPPA QVSVPFMSPA SAYQWFYDGY PTFGEHKQEK DLEYGACPNN MMGTFSVRTV GS SKSKYAL VVRIYMRMKH VRAWIPRPMR NQNYLFKANP NYAGDSIKPT GTSRNAITTL UniProtKB: Genome polyprotein |
-Macromolecule #4: SPHINGOSINE
| Macromolecule | Name: SPHINGOSINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: SPH |
|---|---|
| Molecular weight | Theoretical: 299.492 Da |
| Chemical component information |  ChemComp-SPH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 2739 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)