[English] 日本語
 Yorodumi
Yorodumi- EMDB-50157: Tau PHF subtomogram average relating to CS6 extended data Figure 9e -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tau PHF subtomogram average relating to CS6 extended data Figure 9e | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / Filament / Alzheimer's / helical / PROTEIN FIBRIL | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.0 Å | |||||||||
 Authors Authors | Jenkins J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: CryoET of β-amyloid and tau within postmortem Alzheimer's disease brain. Authors: Madeleine A G Gilbert / Nayab Fatima / Joshua Jenkins / Thomas J O'Sullivan / Andreas Schertel / Yehuda Halfon / Martin Wilkinson / Tjado H J Morrema / Mirjam Geibel / Randy J Read / Neil A ...Authors: Madeleine A G Gilbert / Nayab Fatima / Joshua Jenkins / Thomas J O'Sullivan / Andreas Schertel / Yehuda Halfon / Martin Wilkinson / Tjado H J Morrema / Mirjam Geibel / Randy J Read / Neil A Ranson / Sheena E Radford / Jeroen J M Hoozemans / René A W Frank /    Abstract: A defining pathological feature of most neurodegenerative diseases is the assembly of proteins into amyloid that form disease-specific structures. In Alzheimer's disease, this is characterized by the ...A defining pathological feature of most neurodegenerative diseases is the assembly of proteins into amyloid that form disease-specific structures. In Alzheimer's disease, this is characterized by the deposition of β-amyloid and tau with disease-specific conformations. The in situ structure of amyloid in the human brain is unknown. Here, using cryo-fluorescence microscopy-targeted cryo-sectioning, cryo-focused ion beam-scanning electron microscopy lift-out and cryo-electron tomography, we determined in-tissue architectures of β-amyloid and tau pathology in a postmortem Alzheimer's disease donor brain. β-amyloid plaques contained a mixture of fibrils, some of which were branched, and protofilaments, arranged in parallel arrays and lattice-like structures. Extracellular vesicles and cuboidal particles defined the non-amyloid constituents of β-amyloid plaques. By contrast, tau inclusions formed parallel clusters of unbranched filaments. Subtomogram averaging a cluster of 136 tau filaments in a single tomogram revealed the polypeptide backbone conformation and filament polarity orientation of paired helical filaments within tissue. Filaments within most clusters were similar to each other, but were different between clusters, showing amyloid heterogeneity that is spatially organized by subcellular location. The in situ structural approaches outlined here for human donor tissues have applications to a broad range of neurodegenerative diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50157.map.gz emd_50157.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50157-v30.xml emd-50157-v30.xml emd-50157.xml emd-50157.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50157_fsc.xml emd_50157_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_50157.png emd_50157.png | 13.1 KB | ||
| Masks |  emd_50157_msk_1.map emd_50157_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50157.cif.gz emd-50157.cif.gz | 5.1 KB | ||
| Others |  emd_50157_half_map_1.map.gz emd_50157_half_map_1.map.gz emd_50157_half_map_2.map.gz emd_50157_half_map_2.map.gz | 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50157 http://ftp.pdbj.org/pub/emdb/structures/EMD-50157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50157 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50157.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50157.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.38 Å | ||||||||||||||||||||||||||||||||||||
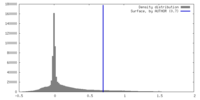
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50157_msk_1.map emd_50157_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
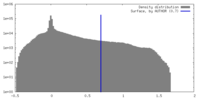
| Density Histograms |
-Half map: #1
| File | emd_50157_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
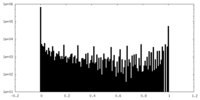
| Density Histograms |
-Half map: #2
| File | emd_50157_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
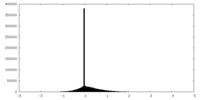
| Density Histograms |
- Sample components
Sample components
-Entire : Microtubule-associated protein tau
| Entire | Name: Microtubule-associated protein tau |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule-associated protein tau
| Supramolecule | Name: Microtubule-associated protein tau / type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Brain Homo sapiens (human) / Tissue: Brain |
-Macromolecule #1: Microtubule-associated protein tau
| Macromolecule | Name: Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKST PTAEDVTAPL VDEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA G HVTQEPES GKVVQEGFLR EPGPPGLSHQ LMSGMPGAPL LPEGPREATR ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKST PTAEDVTAPL VDEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA G HVTQEPES GKVVQEGFLR EPGPPGLSHQ LMSGMPGAPL LPEGPREATR QPSGTGPEDT EG GRHAPEL LKHQLLGDLH QEGPPLKGAG GKERPGSKEE VDEDRDVDES SPQDSPPSKA SPA QDGRPP QTAAREATSI PGFPAEGAIP LPVDFLSKVS TEIPASEPDG PSVGRAKGQD APLE FTFHV EITPNVQKEQ AHSEEHLGRA AFPGAPGEGP EARGPSLGED TKEADLPEPS EKQPA AAPR GKPVSRVPQL KARMVSKSKD GTGSDDKKAK TSTRSSAKTL KNRPCLSPKH PTPGSS DPL IQPSSPAVCP EPPSSPKYVS SVTSRTGSSG AKEMKLKGAD GKTKIATPRG AAPPGQK GQ ANATRIPAKT PPAPKTPPSS GEPPKSGDRS GYSSPGSPGT PGSRSRTPSL PTPPTREP K KVAVVRTPPK SPSSAKSRLQ TAPVPMPDLK NVKSKIGSTE NLKHQPGGGK VQIINKKLD LSNVQSKCGS KDNIKHVPGG GSVQIVYKPV DLSKVTSKCG SLGNIHHKPG GGQVEVKSEK LDFKDRVQS KIGSLDNITH VPGGGNKKIE THKLTFRENA KAKTDHGAEI VYKSPVVSGD T SPRHLSNV SSTGSIDMVD SPQLATLADE VSASLAKQGL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.5 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)