[English] 日本語
 Yorodumi
Yorodumi- EMDB-50149: Myo-inositol-1-phosphate synthase from Thermochaetoides thermophi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | |||||||||||||||
 Map data Map data | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 1 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | inositol metabolism / endogenous / conformational selection / ISOMERASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationinositol-3-phosphate synthase / inositol-3-phosphate synthase activity / inositol biosynthetic process / phospholipid biosynthetic process Similarity search - Function | |||||||||||||||
| Biological species |  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.48 Å | |||||||||||||||
 Authors Authors | Traeger TK / Kyrilis FL / Hamdi F / Kastritis PL | |||||||||||||||
| Funding support | European Union,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway. Authors: Toni K Träger / Fotis L Kyrilis / Farzad Hamdi / Christian Tüting / Marie Alfes / Tommy Hofmann / Carla Schmidt / Panagiotis L Kastritis /   Abstract: Myo-inositol-1-phosphate synthase (MIPS) catalyzes the NAD-dependent isomerization of glucose-6-phosphate (G6P) into inositol-1-phosphate (IMP), controlling the rate-limiting step of the inositol ...Myo-inositol-1-phosphate synthase (MIPS) catalyzes the NAD-dependent isomerization of glucose-6-phosphate (G6P) into inositol-1-phosphate (IMP), controlling the rate-limiting step of the inositol pathway. Previous structural studies focused on the detailed molecular mechanism, neglecting large-scale conformational changes that drive the function of this 240 kDa homotetrameric complex. In this study, we identified the active, endogenous MIPS in cell extracts from the thermophilic fungus . By resolving the native structure at 2.48 Å (FSC = 0.143), we revealed a fully populated active site. Utilizing 3D variability analysis, we uncovered conformational states of MIPS, enabling us to directly visualize an order-to-disorder transition at its catalytic center. An acyclic intermediate of G6P occupied the active site in two out of the three conformational states, indicating a catalytic mechanism where electrostatic stabilization of high-energy intermediates plays a crucial role. Examination of all isomerases with known structures revealed similar fluctuations in secondary structure within their active sites. Based on these findings, we established a conformational selection model that governs substrate binding and eventually inositol availability. In particular, the ground state of MIPS demonstrates structural configurations regardless of substrate binding, a pattern observed across various isomerases. These findings contribute to the understanding of MIPS structure-based function, serving as a template for future studies targeting regulation and potential therapeutic applications. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50149.map.gz emd_50149.map.gz | 483.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50149-v30.xml emd-50149-v30.xml emd-50149.xml emd-50149.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50149_fsc.xml emd_50149_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_50149.png emd_50149.png | 64.8 KB | ||
| Masks |  emd_50149_msk_1.map emd_50149_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50149.cif.gz emd-50149.cif.gz | 6.8 KB | ||
| Others |  emd_50149_additional_1.map.gz emd_50149_additional_1.map.gz emd_50149_additional_2.map.gz emd_50149_additional_2.map.gz emd_50149_half_map_1.map.gz emd_50149_half_map_1.map.gz emd_50149_half_map_2.map.gz emd_50149_half_map_2.map.gz | 483.4 MB 483.8 MB 474.7 MB 474.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50149 http://ftp.pdbj.org/pub/emdb/structures/EMD-50149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50149 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50149 | HTTPS FTP |
-Validation report
| Summary document |  emd_50149_validation.pdf.gz emd_50149_validation.pdf.gz | 985.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50149_full_validation.pdf.gz emd_50149_full_validation.pdf.gz | 985.2 KB | Display | |
| Data in XML |  emd_50149_validation.xml.gz emd_50149_validation.xml.gz | 26.2 KB | Display | |
| Data in CIF |  emd_50149_validation.cif.gz emd_50149_validation.cif.gz | 34.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50149 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50149 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50149 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50149 | HTTPS FTP |
-Related structure data
| Related structure data |  9f2kMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50149.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50149.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.59 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50149_msk_1.map emd_50149_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
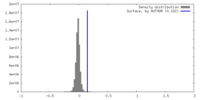
| Density Histograms |
-Additional map: Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 3...
| File | emd_50149_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
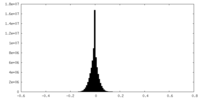
| Density Histograms |
-Additional map: Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 2...
| File | emd_50149_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila; state 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 5; state1
| File | emd_50149_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 5; state1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap A; state1
| File | emd_50149_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap A; state1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Native homotetramer of the Myo-inositol-1-phosphate synthase
| Entire | Name: Native homotetramer of the Myo-inositol-1-phosphate synthase |
|---|---|
| Components |
|
-Supramolecule #1: Native homotetramer of the Myo-inositol-1-phosphate synthase
| Supramolecule | Name: Native homotetramer of the Myo-inositol-1-phosphate synthase type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 240 KDa |
-Macromolecule #1: inositol-3-phosphate synthase
| Macromolecule | Name: inositol-3-phosphate synthase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: inositol-3-phosphate synthase |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) |
| Molecular weight | Theoretical: 56.735477 KDa |
| Sequence | String: IFKVNSPNVV YTDDEIRSKY VYRTTEVTTA EDGSLIATPR ETVYDFKVDR KLPKLGVMLV GWGGNNGSTI TAGIIANRRG LVWETRNGK QEANYYGSVI MGSTIKLGTD AKTHKDINIP FHSVLPMVHP NDIVIGGWDI SGLNLADAMD RAQVLEPSLK A LVRKEMAS ...String: IFKVNSPNVV YTDDEIRSKY VYRTTEVTTA EDGSLIATPR ETVYDFKVDR KLPKLGVMLV GWGGNNGSTI TAGIIANRRG LVWETRNGK QEANYYGSVI MGSTIKLGTD AKTHKDINIP FHSVLPMVHP NDIVIGGWDI SGLNLADAMD RAQVLEPSLK A LVRKEMAS MKPLPSIYYP DFIAANQEDR ADNILPGNKK CWEHVEEIRK NIRDFKAANG LDKVIVLWTA NTERYASIIE GV NDTADNL LNAIKNGHEE VSPSTVFAVS SILEGVPFIN GSPQNTFVPG CIELAERHGA FIGGDDFKSG QTKMKSALVD FLI NAGIKL TSIASYNHLG NNDGKNLSSQ RQFRSKEISK SNVVDDMVEA NTVLYKPGEH PDHIVVIKYV PAVGDSKRAM DEYH GEIFL GGHQTISIAN VCEDSLLASP LIIDLVIVAE LMTRIQWRLH KEDATEADWK YFHSVLSILS YMLKAPMTPP GTPVV NALA KQRAAMANIF RACLGLDPEN DMTLEHKLF UniProtKB: inositol-3-phosphate synthase |
-Macromolecule #2: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 400.0 kPa / Details: 15 mA | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 6 s blot time with a blot force of -1. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.15 K / Max: 103.15 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 2 / Number real images: 6261 / Average electron dose: 28.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 240000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: Local Installation |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9f2k: |
-Atomic model buiding 2
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: Local Installation |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-9f2k: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)