+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the Pyrococcus furiosus SHI complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hydrogen production / [NiFe] hydrogenase / cryo-EM structure / electron transport | |||||||||
| Function / homology |  Function and homology information Function and homology informationsulfhydrogenase / sulfur reductase activity / hydrogen dehydrogenase (NADP+) / hydrogen dehydrogenase (NADP+) activity / [Ni-Fe] hydrogenase complex / pyrimidine nucleotide biosynthetic process / ferredoxin hydrogenase activity / 3 iron, 4 sulfur cluster binding / nickel cation binding / 2 iron, 2 sulfur cluster binding ...sulfhydrogenase / sulfur reductase activity / hydrogen dehydrogenase (NADP+) / hydrogen dehydrogenase (NADP+) activity / [Ni-Fe] hydrogenase complex / pyrimidine nucleotide biosynthetic process / ferredoxin hydrogenase activity / 3 iron, 4 sulfur cluster binding / nickel cation binding / 2 iron, 2 sulfur cluster binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Xiao X / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus. Authors: Xiansha Xiao / Gerrit J Schut / Xiang Feng / Patrick M McTernan / Dominik K Haja / William N Lanzilotta / Michael W W Adams / Huilin Li /  Abstract: The cytoplasmic hydrogenase I (SHI) from the hyperthermophilic archaeon Pyrococcus furiosus belongs to the group III hydrogenase family. SHI oxidizes NADPH rather than NADH to reduce protons and ...The cytoplasmic hydrogenase I (SHI) from the hyperthermophilic archaeon Pyrococcus furiosus belongs to the group III hydrogenase family. SHI oxidizes NADPH rather than NADH to reduce protons and evolve hydrogen gas, and because of this property, coupled with its high thermal stability, the enzyme holds great potential for economical hydrogen production. Despite decades of efforts, the SHI structure has remained unknown. Here, we report the cryoelectron microscopic (cryo-EM) structures of the heterotetrameric SHI holoenzyme (αδβγ). SHI is a symmetric dimer of two individually functional heterotetramers. SHI-αδ resembles the standard [NiFe] hydrogenase, and SHI-βγ function as the NADPH oxidoreductase. SHI-β contains three [4Fe-4S] clusters that relay electrons from NADPH in SHI-γ to the catalytic [NiFe] cluster in SHI-αδ for H production. These structures will guide the adaptation of this unique enzyme for biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_49340.map.gz emd_49340.map.gz | 176.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-49340-v30.xml emd-49340-v30.xml emd-49340.xml emd-49340.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_49340_fsc.xml emd_49340_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_49340.png emd_49340.png | 74.7 KB | ||
| Filedesc metadata |  emd-49340.cif.gz emd-49340.cif.gz | 7.5 KB | ||
| Others |  emd_49340_half_map_1.map.gz emd_49340_half_map_1.map.gz emd_49340_half_map_2.map.gz emd_49340_half_map_2.map.gz | 200 MB 200 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-49340 http://ftp.pdbj.org/pub/emdb/structures/EMD-49340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49340 | HTTPS FTP |
-Validation report
| Summary document |  emd_49340_validation.pdf.gz emd_49340_validation.pdf.gz | 805.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_49340_full_validation.pdf.gz emd_49340_full_validation.pdf.gz | 804.9 KB | Display | |
| Data in XML |  emd_49340_validation.xml.gz emd_49340_validation.xml.gz | 22.1 KB | Display | |
| Data in CIF |  emd_49340_validation.cif.gz emd_49340_validation.cif.gz | 28.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49340 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49340 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49340 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-49340 | HTTPS FTP |
-Related structure data
| Related structure data |  9nezMC  9e15C  9e1jC  9nf0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_49340.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_49340.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_49340_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_49340_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Quaternary complex of the Pfu SHI complex
| Entire | Name: Quaternary complex of the Pfu SHI complex |
|---|---|
| Components |
|
-Supramolecule #1: Quaternary complex of the Pfu SHI complex
| Supramolecule | Name: Quaternary complex of the Pfu SHI complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
-Macromolecule #1: Sulfhydrogenase 1 subunit delta
| Macromolecule | Name: Sulfhydrogenase 1 subunit delta / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: hydrogen dehydrogenase (NADP+) |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 29.269037 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Sequence | String: MGKVRIGFYA LTSCYGCQLQ LAMMDELLQL IPNAEIVCWF MIDRDSIEDE KVDIAFIEGS VSTEEEVELV KKIRENAKIV VAVGACAVQ GGVQSWSEKP LEELWKKVYG DAKVKFQPKK AEPVSKYIKV DYNIYGCPPE KKDFLYALGT FLIGSWPEDI D YPVCLECR ...String: MGKVRIGFYA LTSCYGCQLQ LAMMDELLQL IPNAEIVCWF MIDRDSIEDE KVDIAFIEGS VSTEEEVELV KKIRENAKIV VAVGACAVQ GGVQSWSEKP LEELWKKVYG DAKVKFQPKK AEPVSKYIKV DYNIYGCPPE KKDFLYALGT FLIGSWPEDI D YPVCLECR LNGHPCILLE KGEPCLGPVT RAGCNARCPG FGVACIGCRG AIGYDVAWFD SLAKVFKEKG MTKEEIIERM KM FNGHDER VEKMVEKIFS GGEQ UniProtKB: Sulfhydrogenase 1 subunit delta |
-Macromolecule #2: Sulfhydrogenase 1 subunit beta
| Macromolecule | Name: Sulfhydrogenase 1 subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: sulfhydrogenase |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 43.464688 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Sequence | String: MRYVKLPKEN TYEFLERLKD WGKLYAPVKI SDKFYDFREI DDVRKIEFHY NRTIMPPKKF FFKPREKLFE FDISKPEYRE VIEEVEPFI IFGVHACDIY GLKILDTVYL DEFPDKYYKV RREKGIIIGI SCMPDEYCFC NLRETDFADD GFDLFFHELP D GWLVRVGT ...String: MRYVKLPKEN TYEFLERLKD WGKLYAPVKI SDKFYDFREI DDVRKIEFHY NRTIMPPKKF FFKPREKLFE FDISKPEYRE VIEEVEPFI IFGVHACDIY GLKILDTVYL DEFPDKYYKV RREKGIIIGI SCMPDEYCFC NLRETDFADD GFDLFFHELP D GWLVRVGT PTGHRLVDKN IKLFEEVTDK DICAFRDFEK RRQQAFKYHE DWGNLRYLLE LEMEHPMWDE EADKCLACGI CN TTCPTCR CYEVQDIVNL DGVTGYRERR WDSCQFRSHG LVAGGHNFRP TKKDRFRNRY LCKNAYNEKL GLSYCVGCGR CTA FCPANI SFVGNLRRIL GLEENKCPPT VSEEIPKRGF AYSSNIRGDG V UniProtKB: Sulfhydrogenase 1 subunit beta |
-Macromolecule #3: Sulfhydrogenase 1 subunit gamma
| Macromolecule | Name: Sulfhydrogenase 1 subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: sulfhydrogenase |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 33.093527 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Sequence | String: MMLPKEIMMP NDNPYALHRV KVLKVYSLTE TEKLFLFRFE DPELAEKWTF KPGQFVQLTI PGVGEVPISI CSSPMRKGFF ELCIRKAGR VTTVVHRLKP GDTVLVRGPY GNGFPVDEWE GMDLLLIAAG LGTAPLRSVF LYAMDNRWKY GNITFINTAR Y GKDLLFYK ...String: MMLPKEIMMP NDNPYALHRV KVLKVYSLTE TEKLFLFRFE DPELAEKWTF KPGQFVQLTI PGVGEVPISI CSSPMRKGFF ELCIRKAGR VTTVVHRLKP GDTVLVRGPY GNGFPVDEWE GMDLLLIAAG LGTAPLRSVF LYAMDNRWKY GNITFINTAR Y GKDLLFYK ELEAMKDLAE AENVKIIQSV TRDPNWPGLK GRPQQFIVEA NTNPKNTAVA ICGPPRMYKS VFEALINYGY RP ENIFVTL ERRMKCGIGK CGHCNVGTST SWKYICKDGP VFTYFDIVST PGLLD UniProtKB: Sulfhydrogenase 1 subunit gamma |
-Macromolecule #4: Sulfhydrogenase 1 subunit alpha
| Macromolecule | Name: Sulfhydrogenase 1 subunit alpha / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number: hydrogen dehydrogenase (NADP+) |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 48.381871 KDa |
| Recombinant expression | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Sequence | String: MKNLYLPITI DHIARVEGKG GVEIIIGDDG VKEVKLNIIE GPRFFEAITI GKKLEEALAI YPRICSFCSA AHKLTALEAA EKAVGFVPR EEIQALREVL YIGDMIESHA LHLYLLVLPD YRGYSSPLKM VNEYKREIEI ALKLKNLGTW MMDILGSRAI H QENAVLGG ...String: MKNLYLPITI DHIARVEGKG GVEIIIGDDG VKEVKLNIIE GPRFFEAITI GKKLEEALAI YPRICSFCSA AHKLTALEAA EKAVGFVPR EEIQALREVL YIGDMIESHA LHLYLLVLPD YRGYSSPLKM VNEYKREIEI ALKLKNLGTW MMDILGSRAI H QENAVLGG FGKLPEKSVL EKMKAELREA LPLAEYTFEL FAKLEQYSEV EGPITHLAVK PRGDAYGIYG DYIKASDGEE FP SEKYRDY IKEFVVEHSF AKHSHYKGRP FMVGAISRVI NNADLLYGKA KELYEANKDL LKGTNPFANN LAQALEIVYF IER AIDLLD EALAKWPIKP RDEVEIKDGF GVSTTEAPRG ILVYALKVEN GRVSYADIIT PTAFNLAMME EHVRMMAEKH YNDD PERLK ILAEMVVRAY DPCISCSVHV VRL UniProtKB: Sulfhydrogenase 1 subunit alpha |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 14 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 6 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #7: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 7 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #8: formyl[bis(hydrocyanato-1kappaC)]ironnickel(Fe-Ni)
| Macromolecule | Name: formyl[bis(hydrocyanato-1kappaC)]ironnickel(Fe-Ni) / type: ligand / ID: 8 / Number of copies: 2 / Formula: NFU |
|---|---|
| Molecular weight | Theoretical: 195.591 Da |
| Chemical component information | 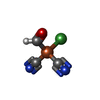 ChemComp-NFU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 17901 / Average exposure time: 1.5 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
| Output model |  PDB-9nez: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































